Parameters and Attributes
Input Parameters
Input parameter |
Description |
|---|---|
data |
(numpy.ndarray) n_samples x n_features. When using via_wrapper(), data is ANNdata object that has a PCA object adata.obsm[‘X_pca’][:, 0:ncomps] and ncomps is the number of components that will be used. |
true_label |
(list) ‘ground truth’ annotations or placeholder |
knn |
(optional, default = 30) number of K-Nearest Neighbors for HNSWlib KNN graph |
root_user |
root_user should be provided as a list containing roots corresponding to index (row number in cell matrix) of root cell. For most trajectories this is of the form [53] where 53 is the index of a sensible root cell, for multiple disconnected trajectories an arbitrary list of cells can be provided [1,506,1100], otherwise VIA arbitratily chooses cells. If the root cells of disconnected trajectories are known in advance, then the cells should be annotated with similar syntax to that of Example Dataset in Disconnected Toy Example 1b. |
dist_std_local |
(optional, default = 1) local pruning threshold for PARC clustering stage: the number of standard deviations above the mean minkowski distance between neighbors of a given node. the higher the parameter, the more edges are retained |
jac_std_global |
(optional, default = 0.15) global level graph pruning for PARC clustering stage. This threshold can also be set as the number of standard deviations below the network’s mean-jaccard-weighted edges. 0.1-1 provide reasonable pruning. higher value means less pruning. e.g. a value of 0.15 means all edges that are above mean(edgeweight)-0.15*std(edge-weights) are retained. We find both 0.15 and ‘median’ to yield good results resulting in pruning away ~ 50-60% edges |
too_big_factor |
(optional, default = 0.4) if a cluster exceeds this share of the entire cell population, then the PARC will be run on the large cluster |
x_lazy |
(optional, default = 0.95) 1-x = probability of staying in same node (lazy). Values between 0.9-0.99 are reasonable |
alpha_teleport |
(optional, default = 0.99) 1-alpha is probability of jumping. Values between 0.95-0.99 are reasonable unless prior knowledge of teleportation |
distance |
(optional, default = ‘l2’ euclidean) ‘ip’,’cosine’ |
random_seed |
(optional, default = 42) The random seed to pass to Leiden |
pseudotime_threshold_TS |
(optional, default = 30) Percentile threshold for potential node to qualify as Terminal State |
resolution_parameter |
(optional, default = 1) Uses ModuliartyVP and RBConfigurationVertexPartition |
preserve_disconnected_after_pruning |
(optional, default = False) Cluster-graph pruning can occasionally cause fragmentation that can be repaired (by setting to True) by retaining select edges. |
cluster_graph_pruning_std |
(optional, default =0.15) Often set to the same value as the PARC clustering level of jac_std_global. To retain more connectivity in the clustergraph underlying the trajectory computations, increase the value |
visual_cluster_graph_pruning |
(optional, default = 0.15) This only comes into play if the user deliberately chooses not to use the default edge-bundling method of visualizating edges (draw_piechart_graph()) and instead calls draw_piechart_graph_nobundle(). It is often set to the same value as the PARC clustering level of jac_std_global. This does not impact computation of terminal states, pseudotime or lineage likelihoods. It controls the number of edges plotted for visual effect |
num_sim_branch_probability |
(optional), default = 500. Number of MCMCs run per terminal state. This can be safely reduced to 100 when computational resources are limited |
small_pop |
(optional, default = 10) Via attempts to merge Clusters with a population < 10 cells with larger clusters. |
edgebundle_pruning |
(optional), default = None. This is automatically set to be the same as cluster_graph_pruning_std |
edgebundle_pruning_twice |
(optional, default = False) If the visualized cluster graph edges seem too busy, they can be further condensed by a second iteration of edge bundling by setting this to True. |
gene_matrix |
(optional, default = None) Only required when using RNA velocity to guide direction. Gene matrix not numpy array: adata.X.todense() |
velocity_matrix |
(optional, default = None). Only required when using RNA velocity to guide direction. Matrix from scVelo with RNA velocities from: adata.layers[‘velocity’] |
velo_weight |
(optional, default = 0.5) #float between 0,1. the weight assigned to directionality and connectivity derived from scRNA-velocity |
pca_loadings |
(optional, default = None) The loadings of the pcs used to project the cells when adjusting the gene-space using velocity: adata.varm[‘PCs’] |
is_coarse |
(optional, default = True) If running VIA in two steps, for the second fine-grained, set to “False’ |
via_coarse |
(optional, default = None) If a second fine-grained iteration of VIA is run using the terminal states, roots and single-cell graph obtained in the first coarse-pass of VIA, then via_coarse = v0 (the VIA object from first iteration) |
Attributes
Attributes |
Description |
|---|---|
labels |
(list) length n_samples of corresponding cluster labels |
single_cell_pt_markov |
(list) computed pseudotime |
single_cell_bp |
(array) computed single cell branch probabilities (lineage likelihoods). n_cells x n_terminal states. The columns each correspond to a terminal state, in the same order presented in the’terminal_clusters’ attribute |
terminal cluster |
(list) terminal clusters found by VIA |
super_cluster_labels |
Set this to v0.labels (clustering output of first pass “v0”) |
super_terminal_cells |
super_terminal_cells = via.get_loc_terminal_states(v0, data) |
full_neighbor_array |
full_neighbor_array = v0.full_neighbor_array. KNN graph from first pass of via - neighbor array |
full_distance_array |
full_distance_array = v0.full_distance_array. KNN graph from first pass of via - edge weights |
ig_full_graph |
ig_full_graph = v0.ig_full_graph igraph of the KNN graph from first pass of via |
csr_array_locally_pruned |
csr_array_locally_pruned = v0.csr_array_locally_pruned. CSR matrix of the locally pruned KNN graph |
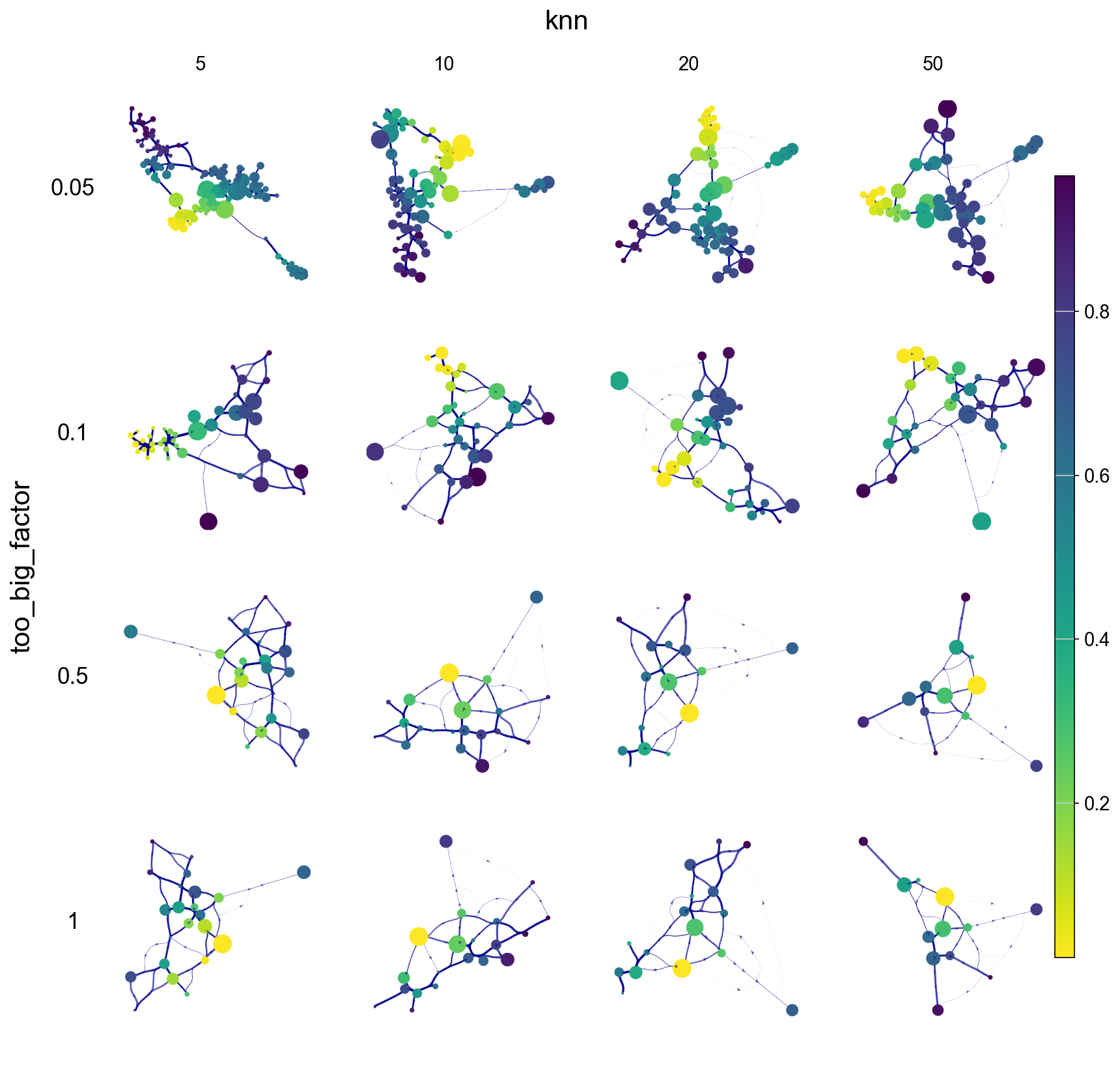
Parameter Effects on VIA cluster-level trajectory graph
knn & too_big_factor effects colored by cell type and pseudotime

jac_std_cluster & cluster_graph_pruning_std effects

