8. StaVia (Via 2.0) for scATAC-seq Hematopoiesis
We show an example of automated cell fate prediction and pathway determination, and visualization for TI, on a scATAC-seq dataset
import pandas as pd
import matplotlib.pyplot as plt
import pyVIA.core as via
import warnings
warnings.filterwarnings('ignore')
#data available on StaVia's github page as a csv file, containing the TFs, cell annotations and Principle Components used in the original paper by Buenrostro et al.,
df = pd.read_csv('/home/user/Trajectory/Datasets/scATAC_Hemato/scATAC_hemato_Buenrostro.csv', sep=',')
print('number cells', df.shape[0])
cell_types = ['GMP', 'HSC', 'MEP', 'CLP', 'CMP', 'LMuPP', 'MPP', 'pDC', 'mono', 'UNK']
cell_dict = {'UNK': 'gray', 'pDC': 'purple', 'mono': 'gold', 'GMP': 'orange', 'MEP': 'red', 'CLP': 'aqua',
'HSC': 'black', 'CMP': 'moccasin', 'MPP': 'darkgreen', 'LMuPP': 'limegreen'}
cell_annot = df['cellname'].values
true_label = []
count = 0
found_annot = False
#reformatting the celltype labels
for annot in cell_annot:
for cell_type_i in cell_types:
if cell_type_i in annot:
true_label.append(cell_type_i)
found_annot = True
if found_annot == False:
true_label.append('unknown')
found_annot = False
PCcol = ['PC1', 'PC2', 'PC3', 'PC4', 'PC5'] #columns in the dataframe containing the Principle Components from the TFs. details of the preprocessing method are described in Buenrostro's paper
number cells 2034
knn = 20
random_seed = 4
X_in = df[PCcol].values #using the PCs provided in the Buenrostro et al., paper
start_ncomp = 0
root = [1200] #the 1200th cell in the input is an HSC cell
v0 = via.VIA(X_in, true_label, edgepruning_clustering_resolution=0.5, edgepruning_clustering_resolution_local=1, knn=knn,
too_big_factor=0.3, root_user=root, dataset='', random_seed=random_seed, memory = 2,
preserve_disconnected=False)
v0.run_VIA()
2024-02-15 17:29:20.222939 Running VIA over input data of 2034 (samples) x 5 (features)
2024-02-15 17:29:20.223006 Knngraph has 20 neighbors
2024-02-15 17:29:22.628608 Finished global pruning of 20-knn graph used for clustering at level of 0.5. Kept 63.6 % of edges.
2024-02-15 17:29:22.643187 Number of connected components used for clustergraph is 1
2024-02-15 17:29:22.751012 Commencing community detection
2024-02-15 17:29:22.785685 Finished running Leiden algorithm. Found 31 clusters.
2024-02-15 17:29:22.787772 Merging 4 very small clusters (<10)
2024-02-15 17:29:22.789018 Finished detecting communities. Found 27 communities
2024-02-15 17:29:22.789580 Making cluster graph. Global cluster graph pruning level: 0.15
2024-02-15 17:29:22.797542 Graph has 1 connected components before pruning
2024-02-15 17:29:22.800130 Graph has 5 connected components after pruning
2024-02-15 17:29:22.805491 Graph has 1 connected components after reconnecting
2024-02-15 17:29:22.806314 0.0% links trimmed from local pruning relative to start
2024-02-15 17:29:22.806351 52.4% links trimmed from global pruning relative to start
2024-02-15 17:29:22.810292 component number 0 out of [0]
2024-02-15 17:29:22.816664 The root index, 1200 provided by the user belongs to cluster number 21 and corresponds to cell type HSC
2024-02-15 17:29:22.820309 Computing lazy-teleporting expected hitting times
2024-02-15 17:29:23.687191 ended all multiprocesses, will retrieve and reshape
try rw2 hitting times setup
start computing walks with rw2 method
g.indptr.size, 28
memory for rw2 hittings times 2. Using rw2 based pt
do scaling of pt
2024-02-15 17:29:30.199701 Identifying terminal clusters corresponding to unique lineages...
2024-02-15 17:29:30.199736 Closeness:[6, 15, 17, 20, 22, 23, 24, 25, 26]
2024-02-15 17:29:30.199751 Betweenness:[0, 1, 5, 6, 11, 15, 16, 17, 20, 21, 22, 23, 24, 25, 26]
2024-02-15 17:29:30.199766 Out Degree:[5, 6, 7, 11, 13, 15, 17, 18, 20, 24, 25, 26]
2024-02-15 17:29:30.200347 Terminal clusters corresponding to unique lineages in this component are [5, 6, 11, 15, 17, 20, 22, 23, 24, 25, 26]
TESTING rw2_lineage probability at memory 2
testing rw2 lineage probability at memory 2
g.indptr.size, 28
2024-02-15 17:29:36.281404 Cluster or terminal cell fate 5 is reached 657.0 times
2024-02-15 17:29:36.377637 Cluster or terminal cell fate 6 is reached 598.0 times
2024-02-15 17:29:36.468722 Cluster or terminal cell fate 11 is reached 224.0 times
2024-02-15 17:29:36.576330 Cluster or terminal cell fate 15 is reached 146.0 times
2024-02-15 17:29:36.674248 Cluster or terminal cell fate 17 is reached 90.0 times
2024-02-15 17:29:36.746055 Cluster or terminal cell fate 20 is reached 596.0 times
2024-02-15 17:29:36.842464 Cluster or terminal cell fate 22 is reached 142.0 times
2024-02-15 17:29:36.947835 Cluster or terminal cell fate 23 is reached 134.0 times
2024-02-15 17:29:37.057720 Cluster or terminal cell fate 24 is reached 119.0 times
2024-02-15 17:29:37.165999 Cluster or terminal cell fate 25 is reached 121.0 times
2024-02-15 17:29:37.288452 Cluster or terminal cell fate 26 is reached 119.0 times
2024-02-15 17:29:37.315628 There are (11) terminal clusters corresponding to unique lineages {5: 'CMP', 6: 'MEP', 11: 'GMP', 15: 'pDC', 17: 'mono', 20: 'MEP', 22: 'pDC', 23: 'pDC', 24: 'CLP', 25: 'CLP', 26: 'pDC'}
2024-02-15 17:29:37.315691 Begin projection of pseudotime and lineage likelihood
2024-02-15 17:29:37.578716 Cluster graph layout based on forward biasing
2024-02-15 17:29:37.582665 Starting make edgebundle viagraph...
2024-02-15 17:29:37.582696 Make via clustergraph edgebundle
2024-02-15 17:29:37.832212 Hammer dims: Nodes shape: (27, 2) Edges shape: (64, 3)
2024-02-15 17:29:37.834641 Graph has 1 connected components before pruning
2024-02-15 17:29:37.838025 Graph has 9 connected components after pruning
2024-02-15 17:29:37.850265 Graph has 1 connected components after reconnecting
2024-02-15 17:29:37.851195 23.3% links trimmed from local pruning relative to start
2024-02-15 17:29:37.851239 45.0% links trimmed from global pruning relative to start
2024-02-15 17:29:37.863579 Time elapsed 17.3 seconds
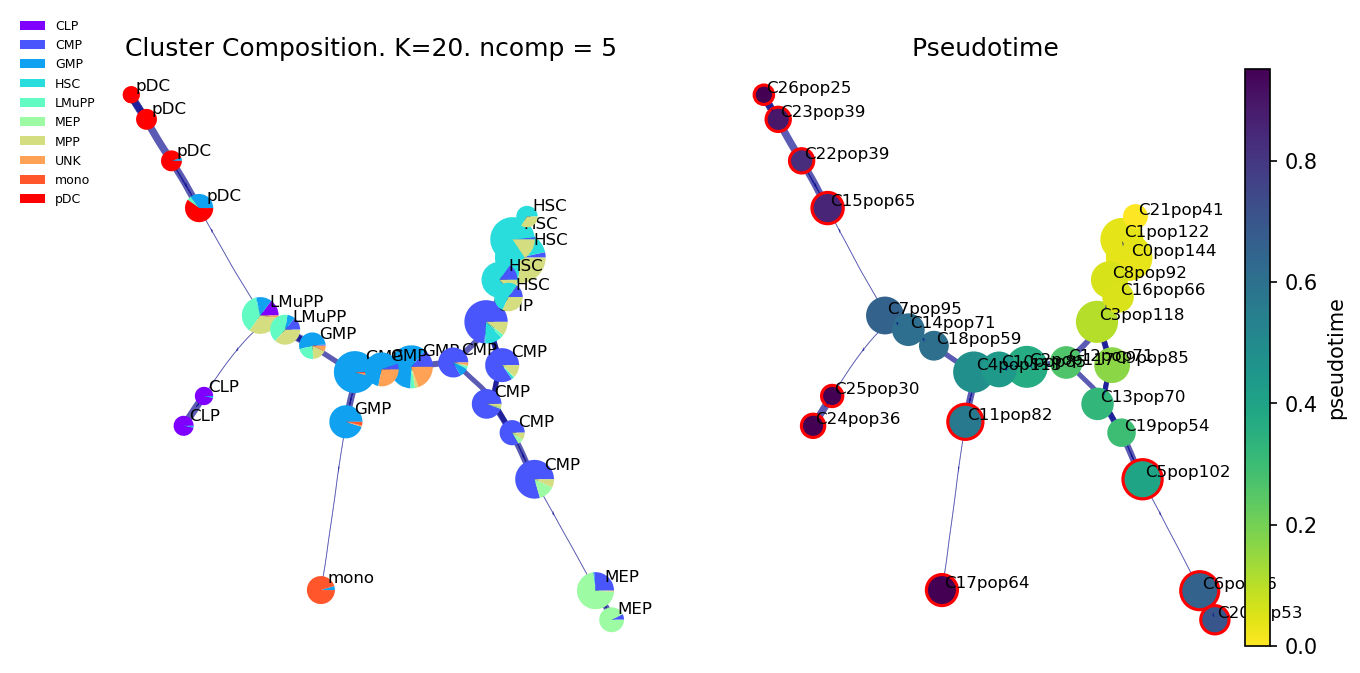
f,ax1, axs2 = via.plot_piechart_viagraph(v0)
f.set_size_inches(10,5)
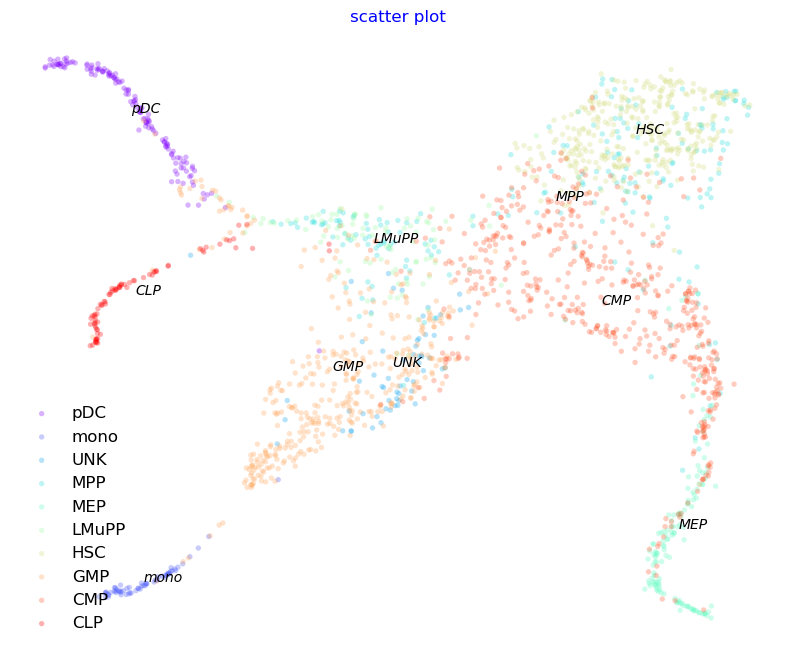
#Create 2D StaVia embedding using the underlying TI infused graph. The 2D embedding will be visually well aligned with the cluster-graph of the trajectory
embedding = via.via_atlas_emb(via_object=v0, min_dist=0.4) #StaVia's TI initialized embedding. Feel free to use a t-SNE, UMAP etc instead
f, ax = via.plot_scatter(embedding=embedding, labels=true_label, s=15)
f.set_size_inches(10,8)
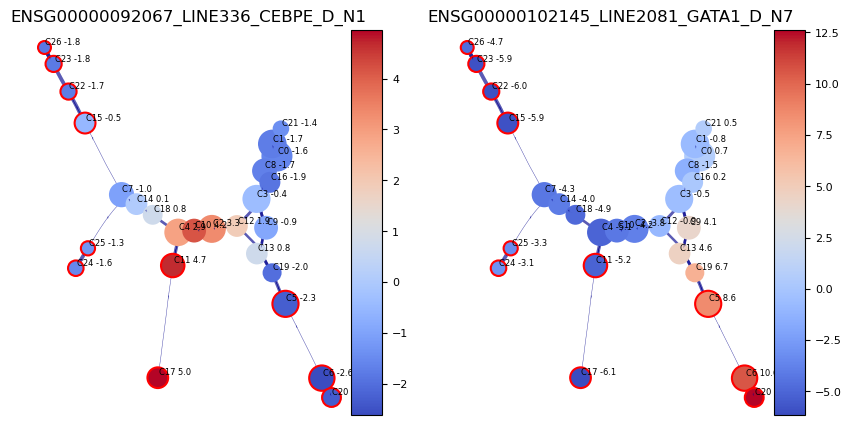
#plot the gene expression of selected genes along the clustergraph
df =df.drop(['cellname'], axis=1)
gene_dict = { 'ENSG00000092067_LINE336_CEBPE_D_N1': 'CEBPE Eosophil (GMP/Mono)','ENSG00000102145_LINE2081_GATA1_D_N7':'GATA1 (MEP)'}
genes_to_plot_list = ['ENSG00000092067_LINE336_CEBPE_D_N1','ENSG00000102145_LINE2081_GATA1_D_N7']
f, axs = via.plot_viagraph( via_object=v0,df_genes=df, gene_list=genes_to_plot_list)
f.set_size_inches(10,5)
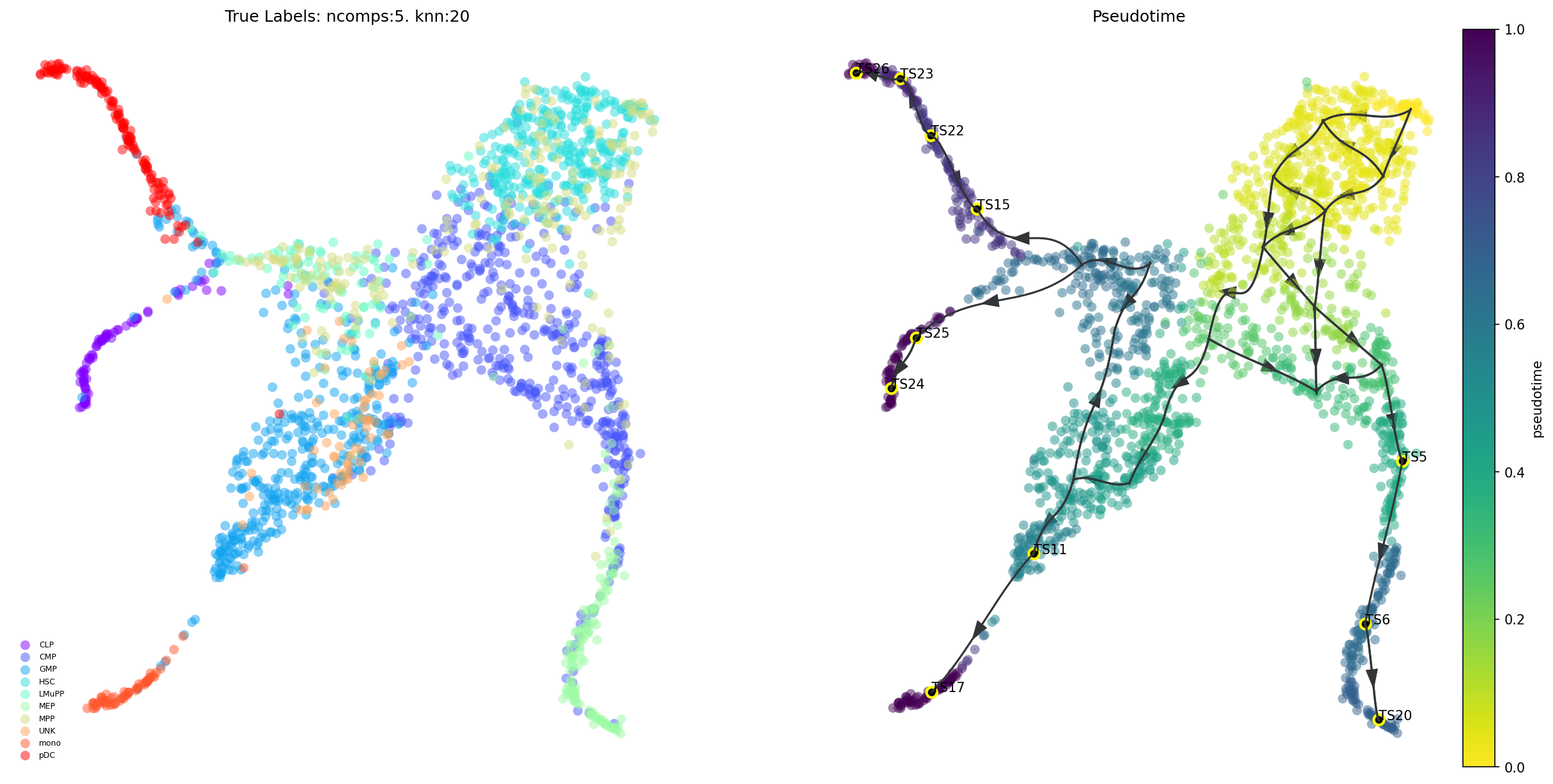
# draw overall pseudotime and main trajectories
via.plot_trajectory_curves(via_object = v0, embedding = embedding,
title_str='Pseudotime')
# draw trajectory and evolution probability for each lineage
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:01 Time: 0:00:01
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:01 Time: 0:00:01
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
2024-02-15 17:35:32.244532 Super cluster 5 is a super terminal with sub_terminal cluster 5
2024-02-15 17:35:32.244619 Super cluster 6 is a super terminal with sub_terminal cluster 6
2024-02-15 17:35:32.244658 Super cluster 11 is a super terminal with sub_terminal cluster 11
2024-02-15 17:35:32.244690 Super cluster 15 is a super terminal with sub_terminal cluster 15
2024-02-15 17:35:32.244719 Super cluster 17 is a super terminal with sub_terminal cluster 17
2024-02-15 17:35:32.244749 Super cluster 20 is a super terminal with sub_terminal cluster 20
2024-02-15 17:35:32.244776 Super cluster 22 is a super terminal with sub_terminal cluster 22
2024-02-15 17:35:32.244808 Super cluster 23 is a super terminal with sub_terminal cluster 23
2024-02-15 17:35:32.244834 Super cluster 24 is a super terminal with sub_terminal cluster 24
2024-02-15 17:35:32.244861 Super cluster 25 is a super terminal with sub_terminal cluster 25
2024-02-15 17:35:32.244886 Super cluster 26 is a super terminal with sub_terminal cluster 26
(<Figure size 3000x1500 with 3 Axes>,
<Axes: title={'center': 'True Labels: ncomps:5. knn:20'}>,
<Axes: title={'center': 'Pseudotime'}>)
via.plot_sc_lineage_probability(via_object=v0, embedding = embedding,marker_lineages=[20,17,24,26])
plt.show()
2024-02-15 17:35:48.454355 Marker_lineages: [20, 17, 24, 26]
2024-02-15 17:35:48.457705 The number of components in the original full graph is 1
2024-02-15 17:35:48.457755 For downstream visualization purposes we are also constructing a low knn-graph
2024-02-15 17:35:50.070895 Check sc pb 0.9999999999999999
f getting majority comp
2024-02-15 17:35:50.197961 Cluster path on clustergraph starting from Root Cluster 21 to Terminal Cluster 5: [21, 1, 8, 3, 9, 19, 5]
2024-02-15 17:35:50.198004 Cluster path on clustergraph starting from Root Cluster 21 to Terminal Cluster 6: [21, 1, 8, 3, 9, 19, 5, 6]
2024-02-15 17:35:50.198030 Cluster path on clustergraph starting from Root Cluster 21 to Terminal Cluster 11: [21, 1, 8, 3, 12, 2, 10, 4, 11]
2024-02-15 17:35:50.198055 Cluster path on clustergraph starting from Root Cluster 21 to Terminal Cluster 15: [21, 1, 8, 3, 12, 2, 10, 4, 18, 14, 7, 15]
2024-02-15 17:35:50.198081 Cluster path on clustergraph starting from Root Cluster 21 to Terminal Cluster 17: [21, 1, 8, 3, 12, 2, 10, 4, 11, 17]
2024-02-15 17:35:50.198106 Cluster path on clustergraph starting from Root Cluster 21 to Terminal Cluster 20: [21, 1, 8, 3, 9, 19, 5, 6, 20]
2024-02-15 17:35:50.198131 Cluster path on clustergraph starting from Root Cluster 21 to Terminal Cluster 22: [21, 1, 8, 3, 12, 2, 10, 4, 18, 14, 7, 15, 22]
2024-02-15 17:35:50.198157 Cluster path on clustergraph starting from Root Cluster 21 to Terminal Cluster 23: [21, 1, 8, 3, 12, 2, 10, 4, 18, 14, 7, 15, 22, 23]
2024-02-15 17:35:50.198182 Cluster path on clustergraph starting from Root Cluster 21 to Terminal Cluster 24: [21, 1, 8, 3, 12, 2, 10, 4, 18, 14, 7, 25, 24]
2024-02-15 17:35:50.198206 Cluster path on clustergraph starting from Root Cluster 21 to Terminal Cluster 25: [21, 1, 8, 3, 12, 2, 10, 4, 18, 14, 7, 25]
2024-02-15 17:35:50.198232 Cluster path on clustergraph starting from Root Cluster 21 to Terminal Cluster 26: [21, 1, 8, 3, 12, 2, 10, 4, 18, 14, 7, 15, 22, 23, 26]
2024-02-15 17:35:50.354424 Revised Cluster level path on sc-knnGraph from Root Cluster 21 to Terminal Cluster 20 along path: [21, 0, 13, 6, 20, 20, 20, 20, 20]
2024-02-15 17:35:50.397034 Revised Cluster level path on sc-knnGraph from Root Cluster 21 to Terminal Cluster 17 along path: [21, 0, 12, 10, 11, 17, 17, 17, 17, 17]
2024-02-15 17:35:50.438633 Revised Cluster level path on sc-knnGraph from Root Cluster 21 to Terminal Cluster 24 along path: [21, 21, 21, 1, 7, 25, 24, 24, 24]
2024-02-15 17:35:50.480060 Revised Cluster level path on sc-knnGraph from Root Cluster 21 to Terminal Cluster 26 along path: [21, 21, 21, 1, 7, 15, 23, 26, 26]
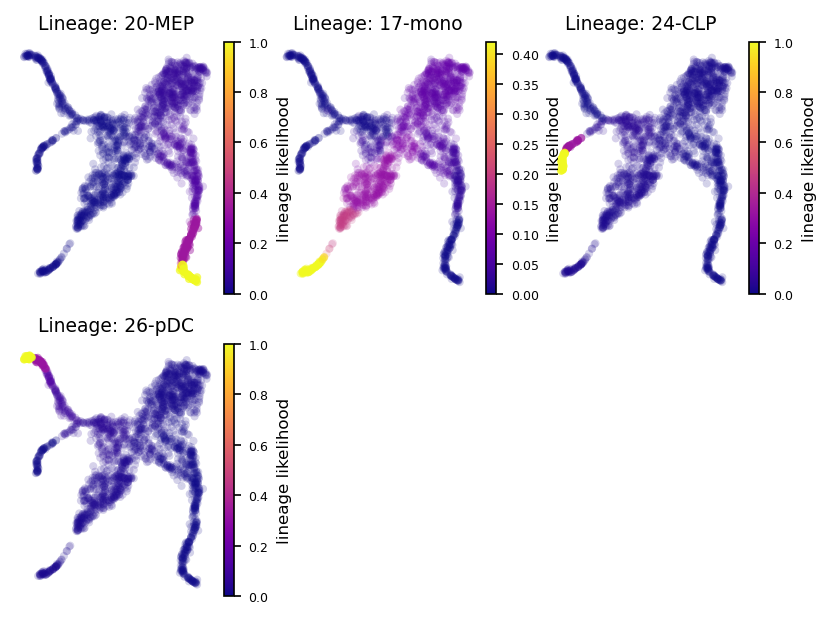
v0.embedding = embedding
f,ax = via.plot_atlas_view(via_object=v0, n_milestones=200, add_sc_embedding=True, sc_labels_expression=v0.single_cell_pt_markov, headwidth_bundle=0.25, linewidth_bundle=2, size_milestones=1, sc_scatter_size=3)
2024-02-15 17:36:03.802544 Computing Edges
2024-02-15 17:36:03.802679 Start finding milestones
2024-02-15 17:36:07.042547 End milestones with 200
2024-02-15 17:36:07.047312 Recompute weights
2024-02-15 17:36:07.069225 pruning milestone graph based on recomputed weights
2024-02-15 17:36:07.070744 Graph has 1 connected components before pruning
2024-02-15 17:36:07.071944 Graph has 1 connected components after pruning
2024-02-15 17:36:07.072208 Graph has 1 connected components after reconnecting
2024-02-15 17:36:07.073549 regenerate igraph on pruned edges
2024-02-15 17:36:07.081077 Setting numeric label as time_series_labels or other sequential metadata for coloring edges
2024-02-15 17:36:07.099631 Making smooth edges
inside add sc embedding second if

#plotting a subset of the detected final cell fates
v0.embedding = embedding
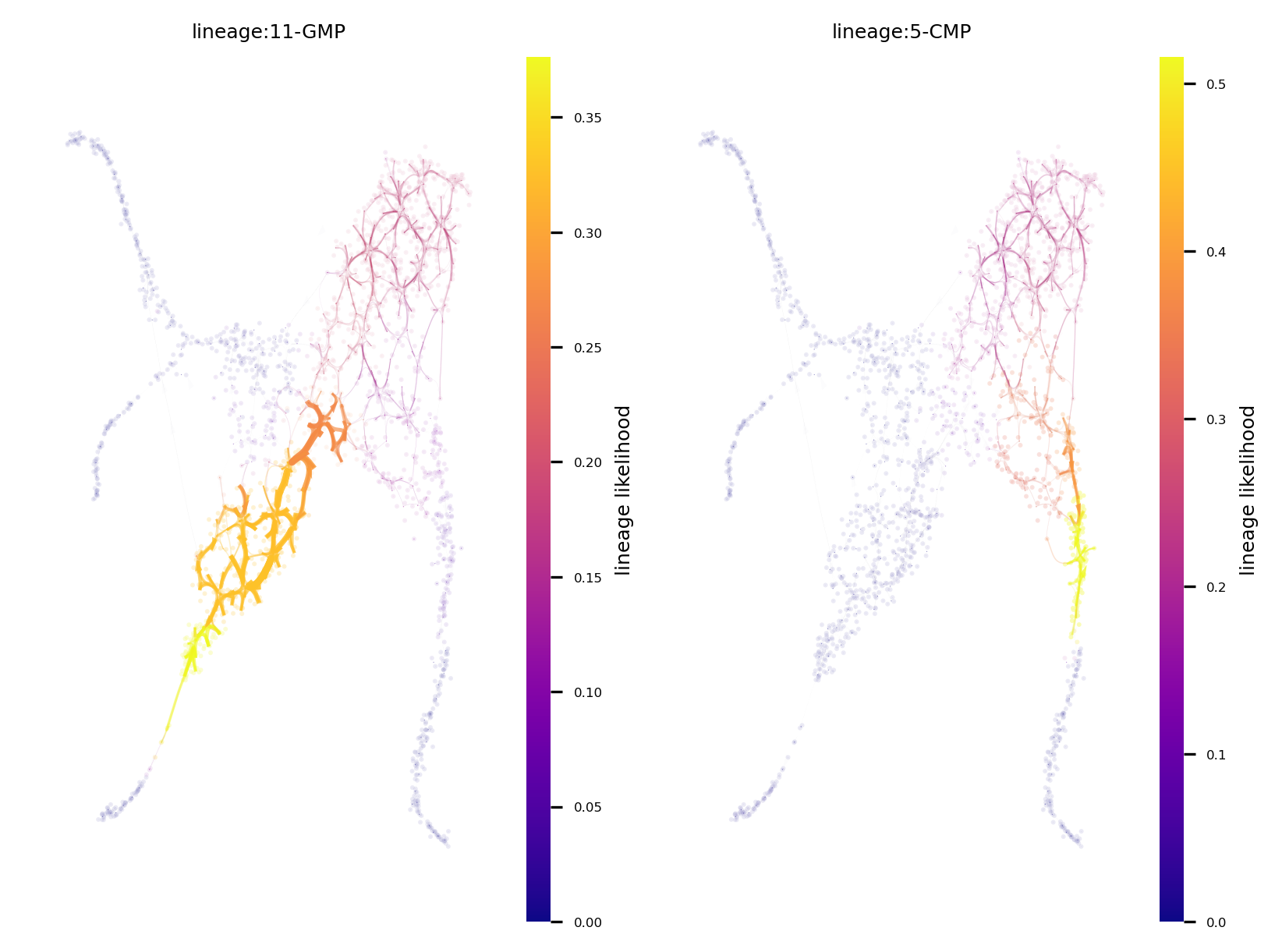
f, ax = via.plot_atlas_view(via_object=v0, n_milestones=400, add_sc_embedding=True, sc_scatter_size=2, size_milestones=0.1,
lineage_pathway=[11,5], headwidth_bundle=0.25, linewidth_bundle=2)
2024-02-15 17:36:25.694978 Computing Edges
2024-02-15 17:36:25.695071 Start finding milestones
2024-02-15 17:36:29.525512 End milestones with 400
2024-02-15 17:36:29.530589 Recompute weights
2024-02-15 17:36:29.568129 pruning milestone graph based on recomputed weights
2024-02-15 17:36:29.569637 Graph has 1 connected components before pruning
2024-02-15 17:36:29.570804 Graph has 1 connected components after pruning
2024-02-15 17:36:29.571118 Graph has 1 connected components after reconnecting
2024-02-15 17:36:29.575342 regenerate igraph on pruned edges
2024-02-15 17:36:29.589088 Setting numeric label as time_series_labels or other sequential metadata for coloring edges
2024-02-15 17:36:29.623256 Making smooth edges
location of 11 is at [2] and 2
location of 5 is at [0] and 0