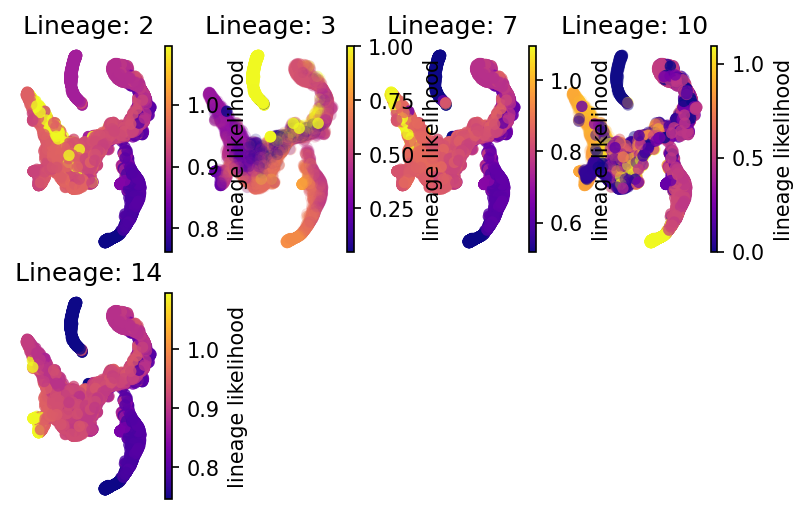7. Bone marrow with RNA-velocity
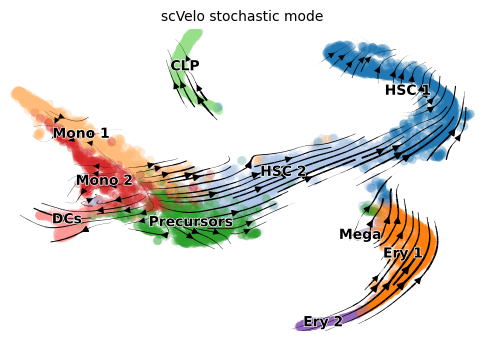
We use the familiar bone marrow dataset (Setty et al 2019) but show how to analyze this dataset using a combination of RNA-velocity and gene-gene similarities. Relying purely on RNA-velocity has been noted to be difficult on this dataset due to a boost in expression (Bergen 2021) which yields negative directionality. By allowing the RNA velocity and gene-gene based graph structure to work together, we can arrive at a more sensible analysis. In Via this is controlled by velo_weight.
Common visualization pitfalls
We also take the opportunity to show that the underlying via graph is less susceptible to directionality artefacts that visualizations that project the trajectory onto umap/tsne/phate for interpretation. There is no doubt that 2D visualizations are useful and intuitive, but we find that can be instances where the visualized directionality is distorted by the 2D single cell embedding and it can therefore be useful to a) refer back to the via graph based abstraction and b) plot the trajectory on a few types of embeddings to overcome any distortions specific to a visualization method.
Load data and pre-process
import core_working as via
import scvelo as scv
import scanpy as sc
import matplotlib.pyplot as plt
adata = scv.datasets.bonemarrow()
print(adata)
n_pcs =80
n_neighbors=30
scv.pp.filter_and_normalize(adata, min_shared_counts=20)#, n_top_genes=hvg)
sc.pp.pca(adata, n_comps=n_pcs)
print('Start scvelo...')
title = 'scVelo stochastic mode' #this is faster but you can also use 'dynamical'
scv.pp.moments(adata, n_pcs=n_pcs, n_neighbors=n_neighbors)
scv.tl.velocity(adata, mode='stochastic')
scv.tl.velocity_graph(adata)
adata.uns['clusters_colors'] = ['#1f77b4', '#aec7e8', '#ff7f0e', '#ffbb78', '#2ca02c', '#98df8a', '#d62728', '#ff9896', '#9467bd', '#c5b0d5']
AnnData object with n_obs × n_vars = 5780 × 14319
obs: 'clusters', 'palantir_pseudotime'
uns: 'clusters_colors'
obsm: 'X_tsne'
layers: 'spliced', 'unspliced'
Filtered out 7837 genes that are detected 20 counts (shared).
Normalized count data: X, spliced, unspliced.
Logarithmized X.
Start scvelo...
computing neighbors
finished (0:00:01) --> added
'distances' and 'connectivities', weighted adjacency matrices (adata.obsp)
computing moments based on connectivities
finished (0:00:03) --> added
'Ms' and 'Mu', moments of un/spliced abundances (adata.layers)
computing velocities
finished (0:00:03) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
computing velocity graph (using 1/8 cores)
0%| | 0/5780 [00:00<?, ?cells/s]
finished (0:00:29) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
The scvelo plot shows negative direction in some regions
scv.pl.velocity_embedding_stream(adata, color='clusters', dpi=100, title=title)
computing velocity embedding
finished (0:00:01) --> added
'velocity_tsne', embedded velocity vectors (adata.obsm)
Run VIA
velocity_matrix = adata.layers['velocity']
gene_matrix = adata.X.todense()
embedding_original = adata.obsm['X_tsne'][:, 0:2]
data_pca = adata.obsm['X_pca'][:, 0:n_pcs]
root = ['HSC_1']
random_seed = 0
jac_std_global = 0.15
cluster_graph_pruning_std=0.3
velo_weight = 0.5 # equal contribution of velocity and gene similarity towards the direction of edges
true_label = [i for i in adata.obs['clusters']]
v0 = via.VIA(data=data_pca, true_label=true_label, jac_std_global=jac_std_global, dist_std_local=1, knn=n_neighbors,
root_user=root, dataset='group', random_seed=random_seed,
is_coarse=True, preserve_disconnected=True, cluster_graph_pruning_std=cluster_graph_pruning_std,
piegraph_arrow_head_width=0.8, piegraph_edgeweight_scalingfactor=2.5, velocity_matrix=velocity_matrix,
gene_matrix=gene_matrix, velo_weight=velo_weight)
v0.run_VIA()
2022-04-29 11:54:57.304416 Running VIA over input data of 5780 (samples) x 80 (features)
2022-04-29 11:54:58.486445 Global pruning of weighted 30 -knn graph
2022-04-29 11:55:00.601603 Finished global pruning. Kept 46.37 of edges.
2022-04-29 11:55:00.633836 Number of connected components used for clustergraph is 1
2022-04-29 11:55:00.936347 the number of components in the original full graph is 1
2022-04-29 11:55:00.936444 For downstream visualization purposes we are also constructing a low knn-graph
2022-04-29 11:55:02.050860 Commencing community detection
2022-04-29 11:55:02.406207 Finished running Leiden algorithm. Found 175 clusters.
2022-04-29 11:55:02.408592 Merging 159 very small clusters (<10)
2022-04-29 11:55:02.415882 Finished detecting communities. Found 16 communities
2022-04-29 11:55:02.417255 Making cluster graph. Global cluster graph pruning level: 0.3
2022-04-29 11:55:02.443449 Graph has 1 connected components before pruning
2022-04-29 11:55:02.444971 Graph has 1 connected components before pruning n_nonz 9 30
2022-04-29 11:55:02.446404 Graph has 1 connected components after reconnecting
2022-04-29 11:55:02.446577 0.0% links trimmed from local pruning relative to start
2022-04-29 11:55:02.446608 57.1% links trimmed from global pruning relative to start
2022-04-29 11:55:03.088915 Looking for initial states
f{datetime.now()} Stationary distribution normed {np.round(pi,3)}
2022-04-29 11:55:03.090192 Top 3 candidates for root: [ 3 10 14] with stationary prob (%) [0.03 2.47 2.83]
2022-04-29 11:55:03.090715 top 5 candidates for terminal: [ 4 8 1 12 9]
new root is 0 with degree 6.16 HSC_1
2022-04-29 11:55:03.152403 Computing lazy-teleporting expected hitting times
2022-04-29 11:55:03.928193 Identifying terminal clusters corresponding to unique lineages...
2022-04-29 11:55:03.928420 Closeness:[0, 3, 7, 10]
2022-04-29 11:55:03.928466 Betweenness:[0, 2, 3, 5, 7, 10, 14]
2022-04-29 11:55:03.928496 Out Degree:[0, 2, 3, 7, 10, 14, 15]
2022-04-29 11:55:03.930588 Terminal clusters corresponding to unique lineages in this component are [2, 3, 7, 10, 14]
2022-04-29 11:55:04.296802 From root 0, the Terminal state 2 is reached 425 times.
2022-04-29 11:55:04.769583 From root 0, the Terminal state 3 is reached 7 times.
2022-04-29 11:55:05.206542 From root 0, the Terminal state 7 is reached 436 times.
2022-04-29 11:55:05.667292 From root 0, the Terminal state 10 is reached 139 times.
2022-04-29 11:55:06.063376 From root 0, the Terminal state 14 is reached 410 times.
2022-04-29 11:55:06.116335 Terminal clusters corresponding to unique lineages are [2, 3, 7, 10, 14]
2022-04-29 11:55:06.116470 Begin projection of pseudotime and lineage likelihood
2022-04-29 11:55:07.245138 Transition matrix with weight of 0.5 on RNA velocity
2022-04-29 11:55:07.246445 Graph has 1 connected components before pruning
2022-04-29 11:55:07.247732 Graph has 1 connected components before pruning n_nonz 2 2
2022-04-29 11:55:07.251005 Graph has 1 connected components after reconnecting
2022-04-29 11:55:07.251188 55.9% links trimmed from local pruning relative to start
2022-04-29 11:55:07.251223 75.0% links trimmed from global pruning relative to start
time elapsed 9.7 seconds
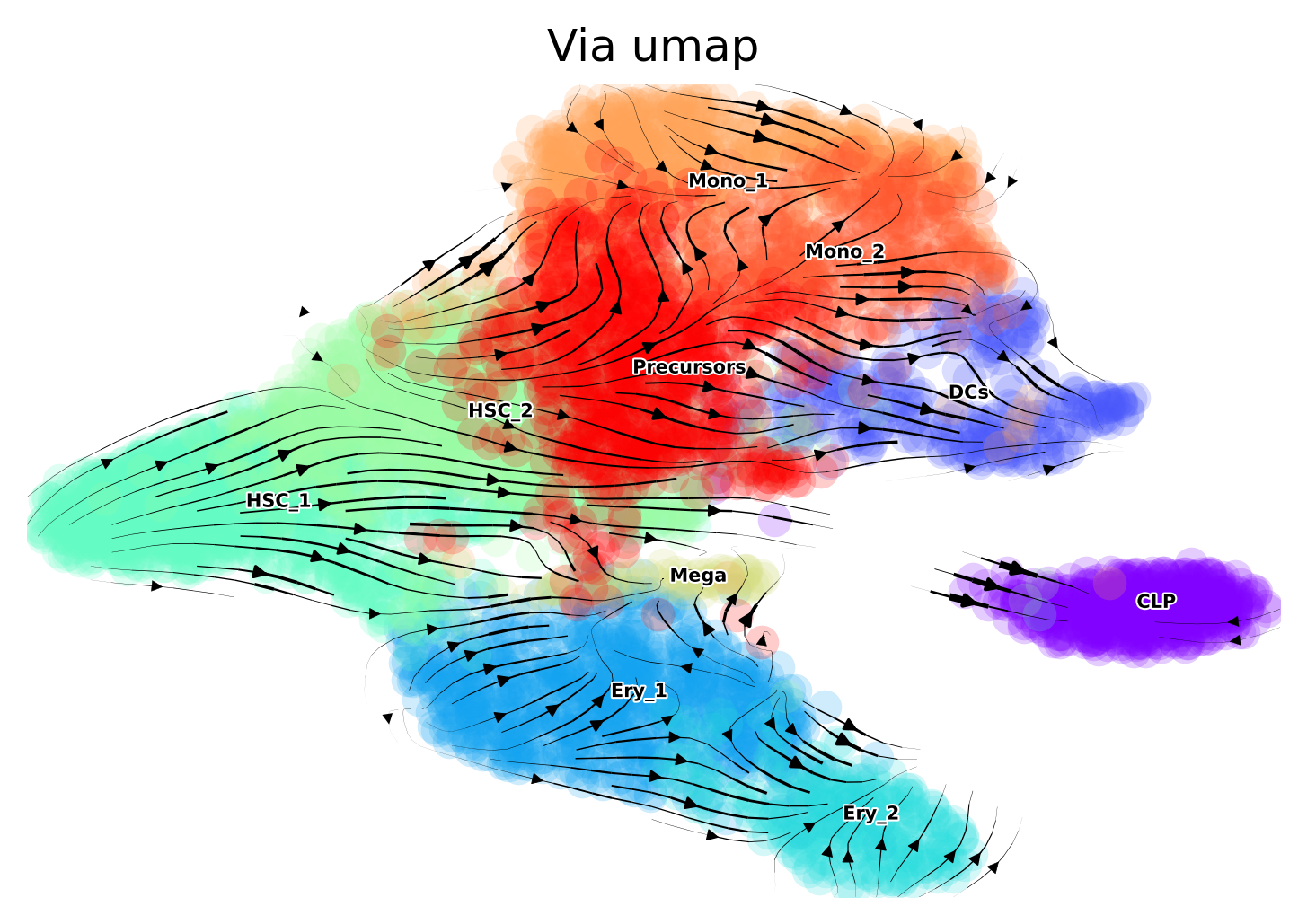
Different embeddings
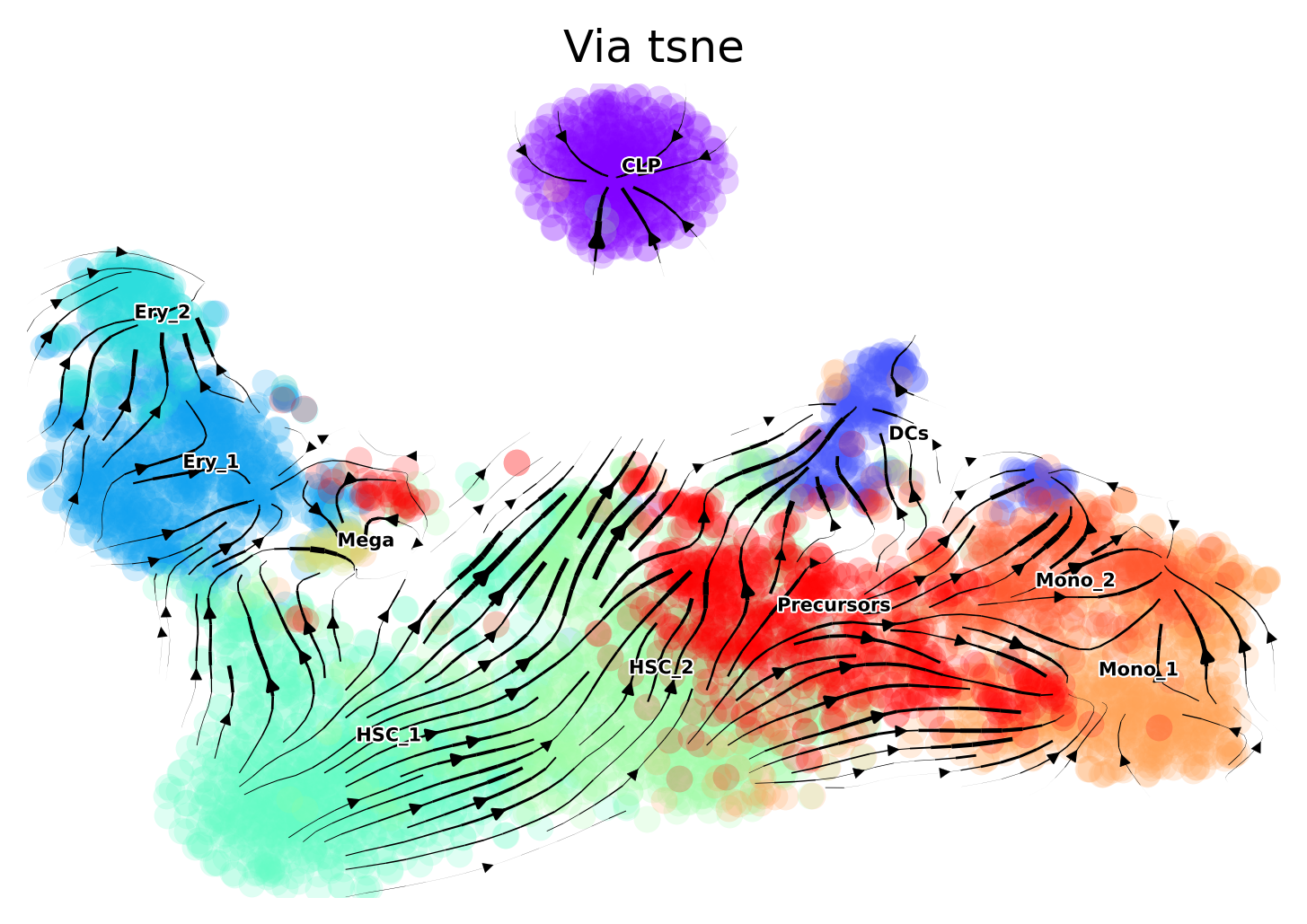
The directionality offered by the Via graph is consistent with the biology. However, when we project the trajectory onto different types of embeddings, we can sometimes see slightly misleading directions. We therefore show the fine-grained vector field of differentiation on the original tsne computed for this dataset and used in publications, a recomputed tsne on scanpy defaults and on umap.
Original tsne You will note that the CLP direction is into itself, however, the HSC2 point in the direction up towards CLP as we expect. Note that in the viagraph, the HSC2s clearly point up to the CLPS. Later we plot the finegrained vector field onto scanpy’s default tsne and umap and see that the overall directinoality is largely consistent with our expectations and the Via graph. This highlights the importance of trying out a few parameters when choosing a suitable embedding.
via.via_streamplot(via_coarse=v0, embedding=embedding_original, scatter_size=50, scatter_alpha=0.2,
marker_edgewidth=0.05, density_stream=2.0, density_grid=1, smooth_transition=2,
smooth_grid=0.5, color_scheme='annotation', add_outline_clusters=False,
cluster_outline_edgewidth=0.001, title='Via original tsne')
plt.show()

Recompute tsne The less of a gap between the CLP cluster and the main body of cells, the less of a misinterpreted arrow we have at the CLP.
print('recompute tsne...')
sc.tl.tsne(adata)
embedding_tsne = adata.obsm['X_tsne']
via.via_streamplot(via_coarse=v0, embedding=embedding_tsne, scatter_size=50, scatter_alpha=0.2,
marker_edgewidth=0.05, density_stream=2.0, density_grid=1, smooth_transition=2,
smooth_grid=0.5, color_scheme='annotation', add_outline_clusters=False,
cluster_outline_edgewidth=0.001, title='Via tsne')
plt.show()
recompute tsne...
Plot on Umap Use scanpy’s default umap implementation. Vector field of trajectory colored by cell type and pseudotime
sc.tl.umap(adata, min_dist=0.8)
embedding_umap = adata.obsm['X_umap']
via.via_streamplot(via_coarse=v0, embedding=embedding_umap, scatter_size=80, scatter_alpha=0.2,
marker_edgewidth=0.05, density_stream=2.0, density_grid=1, smooth_transition=1,
smooth_grid=0.5, color_scheme='annotation', add_outline_clusters=False,
cluster_outline_edgewidth=0.001, title='Via umap')
plt.show()
via.via_streamplot(via_coarse=v0, embedding=embedding_umap, scatter_size=80, scatter_alpha=0.2,
marker_edgewidth=0.05, density_stream=2.0, density_grid=1, smooth_transition=1,
smooth_grid=0.5, color_scheme='time', add_outline_clusters=False,
cluster_outline_edgewidth=0.001, title='Via umap')
plt.show()
graph.data.size 285080
graph.data.size 284804 284804

Lineage Paths
See the differentiation likelihood from the HSC-1 towards each of the detected final states (outlined in Red as detected by VIA)
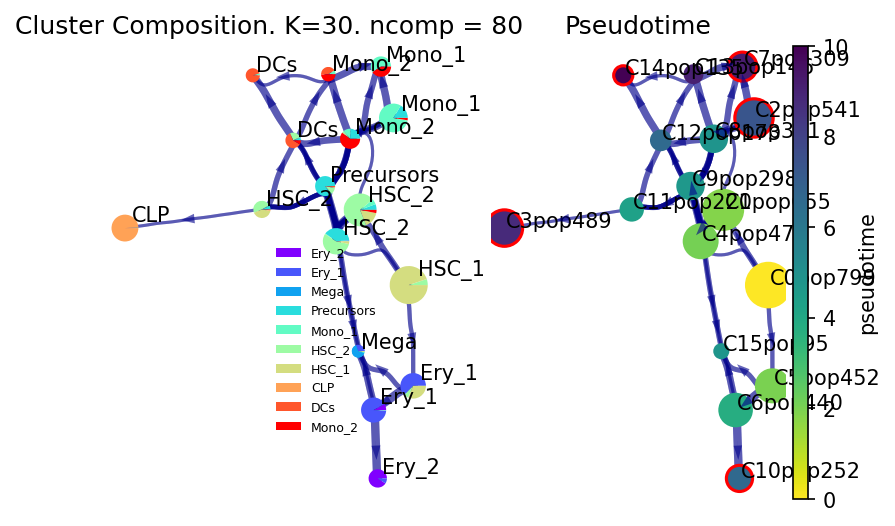
v0.draw_piechart_graph()
via.draw_sc_lineage_probability(v0,v0, embedding=embedding)
plt.show()

Cluster path on clustergraph starting from Root Cluster 0 to Terminal Cluster 2 : [0, 1, 8, 2]
Cluster path on clustergraph starting from Root Cluster 0 to Terminal Cluster 3 : [0, 1, 8, 12, 11, 3]
Cluster path on clustergraph starting from Root Cluster 0 to Terminal Cluster 7 : [0, 1, 8, 2, 7]
Cluster path on clustergraph starting from Root Cluster 0 to Terminal Cluster 10 : [0, 1, 4, 15, 6, 10]
Cluster path on clustergraph starting from Root Cluster 0 to Terminal Cluster 14 : [0, 1, 8, 12, 14]
2022-04-29 11:57:04.548609 Cluster level path on sc-knnGraph from Root Cluster 0 to Terminal Cluster 1 along path: [5, 5, 4, 4, 4, 4, 4, 4, 4]
2022-04-29 11:57:04.572205 Cluster level path on sc-knnGraph from Root Cluster 0 to Terminal Cluster 2 along path: [5, 5, 4, 8, 2, 2, 2]
2022-04-29 11:57:04.593602 Cluster level path on sc-knnGraph from Root Cluster 0 to Terminal Cluster 3 along path: [5, 5, 11, 3, 3, 3, 3]
2022-04-29 11:57:04.615902 Cluster level path on sc-knnGraph from Root Cluster 0 to Terminal Cluster 4 along path: [5, 5, 5, 1, 1, 1, 1]
2022-04-29 11:57:04.637048 Cluster level path on sc-knnGraph from Root Cluster 0 to Terminal Cluster 5 along path: [5, 5, 5, 5, 5]