2. Via 2.0 Cartography on Zebrahub (Trajectory Inference)
This tutorial focuses on how to perform TI analysis of Zebrahub. See Tutorial #3 for Atlas View creation
import scanpy as sc
import matplotlib.pyplot as plt
import numpy as np
import pyVIA.core as via
from datetime import datetime
import pyVIA.datasets_via as datasets
Load the data
Data has been filtered and library normalized and log1p. Data is annotated with time/stage, cluster labels, and cell type labels (coarse and fine).
print(f'{datetime.now()}\tStart reading data')
adata=sc.read_h5ad( filename='/home/user/Trajectory/Datasets/Zebrafish_Lange2023/Rawdata/Zebrahub_data_via.h5ad')
#compute PCA
adata.uns['log1p']['base'] = None #fix for small bug in scanpy #see https://github.com/theislab/single-cell-tutorial/issues/97
sc.pp.highly_variable_genes(adata, n_top_genes=2000)
sc.tl.pca(adata, n_comps=100)
print(adata)
#load the adata_velo below if you want to try to use velocity. this is a 8GB file so is very RAM intensive
adata_velo = sc.read_h5ad('/home/user/Trajectory/Datasets/Zebrafish_Lange2023/Rawdata/Zebrahub_data_velocity_via.h5ad') #10GB file. if you dont have the RAM to load this, then you can still run Via2 without velocity.
print('adata_velo')
print(adata_velo)
#the data can also be loaded using
adata = datasets.zebrahub(foldername="./", use_velocity = False)
2023-09-25 08:25:15.560669 Start reading data
AnnData object with n_obs × n_vars = 119457 × 28187
obs: 'n_genes', 'n_counts', 'fish', 'timepoint_cluster', 'n_genes_by_counts', 'total_counts', 'total_counts_mt', 'pct_counts_mt', 'total_counts_nc', 'pct_counts_nc', 'developmental_stage', 'timepoint', 'zebrafish_anatomy_ontology_class', 'zebrafish_anatomy_ontology_id', 'parc', 'kmeans50', 'celltype_extrainfo', 'zebrafish_anatomy_ontology_class_shobi', 'adjusted_timepoint', 'bool_list', 'global annotation', 'time_coarse_cell_type', 'time_fine_cell_type', 'parc_cluster', 'parc_cluster_and_cell_type'
var: 'gene_ids', 'feature_types', 'genome', 'mt', 'nc', 'n_cells_by_counts', 'mean_counts', 'pct_dropout_by_counts', 'total_counts', 'n_cells', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'global_annotation_colors', 'hvg', 'log1p', 'pca', 'timepoint_colors', 'zebrafish_anatomy_ontology_class_colors', 'zebrafish_anatomy_ontology_id_colors'
obsm: 'X_U', 'X_Via2_Atlas', 'X_pca', 'X_umap'
varm: 'PCs'
layers: 'counts'
adata_velo
AnnData object with n_obs × n_vars = 119457 × 3000
obs: 'clusters', 'seurat_clusters', 'timepoint', 'fish', 'initial_size_spliced', 'initial_size_unspliced', 'initial_size', 'n_counts', 'velocity_self_transition', 'velocity_length', 'velocity_confidence', 'velocity_confidence_transition', 'barcode in labeled data', 'parc', 'kmeans50', 'ontology_class', 'parc_cluster', 'parc_cluster_and_cell_type'
var: 'n_cells', 'highly_variable', 'means', 'dispersions', 'dispersions_norm', 'mean', 'std', 'gene_count_corr', 'velocity_gamma', 'velocity_qreg_ratio', 'velocity_r2', 'velocity_genes', 'fit_alpha', 'fit_beta', 'fit_gamma', 'fit_t_', 'fit_scaling', 'fit_std_u', 'fit_std_s', 'fit_likelihood', 'fit_u0', 'fit_s0', 'fit_pval_steady', 'fit_steady_u', 'fit_steady_s', 'fit_variance', 'fit_alignment_scaling', 'fit_r2'
uns: 'hvg', 'neighbors', 'pca', 'recover_dynamics', 'seurat_clusters_colors', 'umap', 'velocity_graph', 'velocity_graph_neg', 'velocity_params'
obsm: 'X_Via2_Atlas', 'X_pca', 'X_umap', 'noveloumap', 'velocity_umap'
varm: 'PCs', 'loss'
layers: 'Ms', 'Mu', 'counts', 'fit_t', 'fit_tau', 'fit_tau_', 'matrix', 'spliced', 'unspliced', 'variance_velocity', 'velocity', 'velocity_u'
obsp: 'connectivities', 'distances'
Extract some annotation
n_cells = adata.shape[0]
print('cells is', n_cells)
parc_cluster = [i for i in adata.obs['parc_cluster']]
parc_cluster_and_cell_type = [i for i in adata.obs['parc_cluster_and_cell_type']]
coarse_cell_type = [i for i in adata.obs['zebrafish_anatomy_ontology_class']]
fine_cell_type = [i for i in adata.obs['zebrafish_anatomy_ontology_class_shobi']] #consolidated annotaions from individual time point files
coarse_fine_cell_type = [coarse_cell_type[i] + fine_cell_type[i] for i in range(n_cells)]
time_point_numeric = [i for i in adata.obs['adjusted_timepoint']] #assigned a numeric value to each stage (e.g. 5hpf is 5, 10hpf is 10,...)
early_cell_type = [i if str(i) != 'nan' else '' for i in adata.obs['global annotation']]
hybrid_cell_time = [str(fine_cell_type[i]) + '_'+early_cell_type[i]+'_'+str(time_point_numeric[i]) for i in range(n_cells)] #labels of early stage cells which was provided for cells <24hpf also included
parc_numeric_cluster_labels = [i for i in adata.obs['parc']]
parc_cluster = [i for i in adata.obs['parc_cluster']]
cells is 119457
Find a suitable root
root_str = 'paraxial mesoderm_Somites_0'
loc_root = np.where(np.asarray(hybrid_cell_time) == root_str)[0] #'paraxial mesoderm_PSM_0' 1 cell , 'paraxial mesoderm_Somites_0' has 51 cells, 'paraxial mesoderm_Muscle_0', 'tail bud_NMPs_0'
root = [loc_root[0]]
n_pcs = 100
print(f'{datetime.now()}\troot is cell number {root[0]} of type {fine_cell_type[root[0]]}')
2023-09-25 08:28:03.566017 root is cell number 2604 of type paraxial mesoderm
Initialize parameters
knn=30
time_series = True
memory = 50
jac_std_global = 1
random_seed = 0#1
knn_sequential = 5
t_diff_step = 2
do_gaussian_kernel_edgeweights = False
edgebundle_pruning_twice = True
cluster_graph_pruning_std = 0.15
edgebundle_pruning = 0.05
neighboring_terminal_states_threshold = 4
viagraph_decay = 0.7
max_vis_out = 2
use_velocity = True
velo_weight = 0.5
if use_velocity:
gene_matrix =np.asmatrix(adata_velo.layers['Ms'])
velocity_matrix = np.asmatrix(adata_velo.layers['velocity'])
velo_weight=velo_weight
else:
gene_matrix =None
velocity_matrix = None
velo_weight=0
Set terminal states (optional)
# The parameter "user_defined_terminal_group" is being set to a list of group level annotations corresponding to the provided true_label. These were terminal fates detected in previous runs and saved for reproducability.
# This parameter can also be left to its default user_defined_terminal_group = []. the results should be similar.
user_defined_cellfates= ['0_cartilage element', '2_dermis', '49_fin',
'67_pharyngeal arch', '120_cranial cartilage',
'1_forebrain', '71_neuron', '89_diencephalon',
'136_radial glial cell',
'10_epidermal cell', '83_neuromast',
'145_enteric musculature', '55_myotome',
'46_hematopoietic cell', '102_vasculature', '127_Muller cell', '105_oligodendrocyte', '69_liver', '90_pharynx',
'95_intestine']
Initialize and Run the Via2.0 Class
# Initialize and Run the Via2.0 Class
print(f'{datetime.now()}\tRun Via2.0')
v0 = via.VIA(adata.obsm['X_pca'][:, 0:n_pcs], true_label=parc_cluster_and_cell_type, jac_std_global=jac_std_global,
labels=parc_cluster,
dist_std_local=1, knn=knn, memory=memory,
user_defined_terminal_group=user_defined_cellfates,
cluster_graph_pruning_std=cluster_graph_pruning_std,
neighboring_terminal_states_threshold=neighboring_terminal_states_threshold,
too_big_factor=0.3, resolution_parameter=1,
root_user=root, dataset='', random_seed=random_seed,
is_coarse=True, preserve_disconnected=False, pseudotime_threshold_TS=40, x_lazy=0.99,
do_gaussian_kernel_edgeweights=do_gaussian_kernel_edgeweights,
alpha_teleport=0.99, edgebundle_pruning=edgebundle_pruning, edgebundle_pruning_twice=True,
time_series=time_series, time_series_labels=time_point_numeric, knn_sequential=knn_sequential,
knn_sequential_reverse=knn_sequential, t_diff_step=t_diff_step, velo_weight=velo_weight, velocity_matrix=velocity_matrix, gene_matrix=gene_matrix,
do_compute_embedding=False)
v0.run_VIA()
v0.embedding = adata.obsm['X_Via2_Atlas']
2023-09-25 08:28:03.589451 Run Via2.0
2023-09-25 08:28:04.804497 Running VIA over input data of 119457 (samples) x 100 (features)
2023-09-25 08:28:04.804574 Knngraph has 30 neighbors
2023-09-25 08:28:42.727156 Using time series information to guide knn graph construction
2023-09-25 08:28:42.736297 Time series ordered set [0, 5, 10, 15, 20, 25, 30, 35, 40, 45]
2023-09-25 08:29:35.321478 Shape neighbors (119457, 30) and sequential neighbors (119457, 10)
2023-09-25 08:29:35.354718 Shape augmented neighbors (119457, 40)
2023-09-25 08:29:35.380011 Actual average allowable time difference between nodes is 10.0
2023-09-25 08:30:02.089539 Finished global pruning of 30-knn graph used for clustering at level of 1. Kept 87.6 % of edges.
2023-09-25 08:30:02.213207 Number of connected components used for clustergraph is 1
2023-09-25 08:30:18.393414 Using predfined labels provided by user (this must be provided as an array)
2023-09-25 08:30:18.393502 Making cluster graph. Global cluster graph pruning level: 0.15
2023-09-25 08:30:19.415532 Graph has 1 connected components before pruning
2023-09-25 08:30:19.425338 Graph has 1 connected components after pruning
2023-09-25 08:30:19.425605 Graph has 1 connected components after reconnecting
2023-09-25 08:30:19.427311 44.3% links trimmed from local pruning relative to start
2023-09-25 08:30:19.427334 87.4% links trimmed from global pruning relative to start
2023-09-25 08:30:19.430975 Graph has 1 connected components before pruning
2023-09-25 08:30:19.444368 Graph has 3 connected components after pruning
2023-09-25 08:30:19.448015 Graph has 1 connected components after reconnecting
2023-09-25 08:30:19.449751 44.3% links trimmed from local pruning relative to start
2023-09-25 08:30:19.449775 90.1% links trimmed from global pruning relative to start
size velocity matrix 119457 (119457, 3000)
/home/user/anaconda3/envs/Via2Env_py10/lib/python3.10/site-packages/scipy/sparse/_index.py:103: SparseEfficiencyWarning: Changing the sparsity structure of a csr_matrix is expensive. lil_matrix is more efficient.
self._set_intXint(row, col, x.flat[0])
2023-09-25 08:30:23.954500 Looking for initial states
2023-09-25 08:30:24.106221 Stationary distribution normed [0.01 0.005 0.009 0.032 0.015 0.01 0.006 0.006 0.008 0.022 0.008 0.009
0.012 0.007 0.009 0.011 0.006 0.021 0.001 0.002 0.008 0.004 0.003 0.009
0.006 0.014 0.006 0.016 0.011 0.001 0.003 0.008 0.003 0.007 0.007 0.004
0.006 0.001 0.007 0.002 0.005 0.012 0.009 0.005 0.007 0.018 0. 0.005
0.011 0.006 0.007 0.008 0.017 0.006 0.017 0.004 0.005 0.003 0.01 0.005
0.012 0.007 0.015 0.002 0.007 0.005 0.006 0.005 0.018 0.005 0.011 0.006
0. 0.008 0.006 0.006 0.005 0.004 0.003 0.007 0.006 0.008 0.001 0.002
0.011 0.008 0.001 0.005 0.002 0.003 0.005 0.001 0.006 0.003 0.002 0.006
0.006 0.019 0.008 0.007 0.003 0.012 0.001 0.004 0.009 0.001 0.001 0.002
0.012 0.008 0.007 0.011 0.003 0.008 0.005 0.001 0.004 0.006 0.011 0.002
0.005 0.005 0.021 0.009 0.006 0.002 0.005 0.001 0.001 0.002 0.005 0.002
0.008 0.015 0. 0.017 0.006 0.005 0.005 0.004 0.001 0.004 0.006 0.005
0.003 0.005]
2023-09-25 08:30:24.107358 Top 5 candidates for root: [134 72 46 37 82 18 102 127 86 105] with stationary prob (%) [0.032 0.039 0.043 0.055 0.055 0.07 0.076 0.082 0.089 0.092]
2023-09-25 08:30:24.107614 Top 5 candidates for terminal: [ 3 9 122 17 97]
cell type of suggested roots: ['120_cranial cartilage', '24_myotome', '6_ventricular zone', '49_pectoral fin', '89_diencephalon', '74_cranial cartilage', '23_eye', '1_forebrain', '107_neuron', '103_fin']
cell stage of suggested roots: [45, 15, 35, 45, 35, 40, 35, 35, 40, 25]
2023-09-25 08:30:24.122752 component number 0 out of [0]
2023-09-25 08:30:24.460261 The root index, 2604 provided by the user belongs to cluster number 97 and corresponds to cell type 97_paraxial mesoderm
2023-09-25 08:30:24.629263 Computing lazy-teleporting expected hitting times
2023-09-25 08:30:32.855588 ended all multiprocesses, will retrieve and reshape
try rw2 hitting times setup
start computing walks with rw2 method
g.indptr.size, 147
/home/user/anaconda3/envs/Via2Env_py10/lib/python3.10/site-packages/pecanpy/graph.py:90: UserWarning: WARNING: Implicitly set node IDs to the canonical node ordering due to missing IDs field in the raw CSR npz file. This warning message can be suppressed by setting implicit_ids to True in the read_npz function call, or by setting the --implicit_ids flag in the CLI
warnings.warn(
memory for rw2 hittings times 2. Using rw2 based pt
do scaling of pt
2023-09-25 08:30:41.521269 Terminal cluster list based on user defined cells/groups: [('0_cartilage element', 0), ('2_dermis', 2), ('49_fin', 49), ('67_pharyngeal arch', 67), ('120_cranial cartilage', 120), ('1_forebrain', 1), ('71_neuron', 71), ('89_diencephalon', 89), ('136_radial glial cell', 136), ('10_epidermal cell', 10), ('83_neuromast', 83), ('145_enteric musculature', 145), ('55_myotome', 55), ('46_hematopoietic cell', 46), ('102_vasculature', 102), ('127_Muller cell', 127), ('105_oligodendrocyte', 105), ('69_liver', 69), ('90_pharynx', 90), ('95_intestine', 95)]
2023-09-25 08:30:41.521658 Terminal clusters corresponding to unique lineages in this component are [0, 2, 49, 67, 120, 1, 71, 89, 136, 10, 83, 145, 55, 46, 102, 127, 105, 69, 90, 95]
TESTING rw2_lineage probability at memory 50
testing rw2 lineage probability at memory 50
g.indptr.size, 147
/home/user/anaconda3/envs/Via2Env_py10/lib/python3.10/site-packages/pecanpy/graph.py:90: UserWarning: WARNING: Implicitly set node IDs to the canonical node ordering due to missing IDs field in the raw CSR npz file. This warning message can be suppressed by setting implicit_ids to True in the read_npz function call, or by setting the --implicit_ids flag in the CLI
warnings.warn(
2023-09-25 08:30:46.871385 Cluster or terminal cell fate 0 is reached 194.0 times
2023-09-25 08:30:47.265164 Cluster or terminal cell fate 2 is reached 211.0 times
2023-09-25 08:30:47.620006 Cluster or terminal cell fate 49 is reached 38.0 times
2023-09-25 08:30:47.959715 Cluster or terminal cell fate 67 is reached 206.0 times
2023-09-25 08:30:48.442676 Cluster or terminal cell fate 120 is reached 52.0 times
2023-09-25 08:30:48.917698 Cluster or terminal cell fate 1 is reached 265.0 times
2023-09-25 08:30:49.369804 Cluster or terminal cell fate 71 is reached 148.0 times
2023-09-25 08:30:49.691185 Cluster or terminal cell fate 89 is reached 485.0 times
2023-09-25 08:30:50.014253 Cluster or terminal cell fate 136 is reached 137.0 times
2023-09-25 08:30:50.368288 Cluster or terminal cell fate 10 is reached 29.0 times
2023-09-25 08:30:50.585364 Cluster or terminal cell fate 83 is reached 128.0 times
2023-09-25 08:30:50.860216 Cluster or terminal cell fate 145 is reached 37.0 times
2023-09-25 08:30:51.067859 Cluster or terminal cell fate 55 is reached 76.0 times
2023-09-25 08:30:51.453572 Cluster or terminal cell fate 46 is reached 21.0 times
2023-09-25 08:30:51.802014 Cluster or terminal cell fate 102 is reached 62.0 times
2023-09-25 08:30:52.075750 Cluster or terminal cell fate 127 is reached 70.0 times
2023-09-25 08:30:52.443008 Cluster or terminal cell fate 105 is reached 111.0 times
2023-09-25 08:30:52.775031 Cluster or terminal cell fate 69 is reached 26.0 times
2023-09-25 08:30:53.049582 Cluster or terminal cell fate 90 is reached 26.0 times
2023-09-25 08:30:53.420929 Cluster or terminal cell fate 95 is reached 15.0 times
2023-09-25 08:30:54.022736 There are (20) terminal clusters corresponding to unique lineages {0: '0_pectoral fin', 2: '2_dermis', 49: '49_fin', 67: '67_pharyngeal arch', 120: '120_cranial cartilage', 1: '1_forebrain', 71: '71_neuron', 89: '89_diencephalon', 136: '136_radial glial cell', 10: '10_epidermal cell', 83: '83_neuromast', 145: '145_enteric musculature', 55: '55_myotome', 46: '46_hematopoietic cell', 102: '102_vasculature', 127: '127_Muller cell', 105: '105_oligodendrocyte', 69: '69_liver', 90: '90_pharynx', 95: '95_intestine'}
2023-09-25 08:30:54.022861 Begin projection of pseudotime and lineage likelihood
2023-09-25 08:31:03.461077 Start reading data
2023-09-25 08:31:03.461154 Correlation of Via pseudotime with developmental stage 72.11 %
2023-09-25 08:31:03.498392 Transition matrix with weight of 0.5 on RNA velocity
2023-09-25 08:31:03.498789 Cluster graph layout based on forward biasing
2023-09-25 08:31:03.528940 Graph has 1 connected components before pruning
2023-09-25 08:31:03.536532 Graph has 57 connected components after pruning
2023-09-25 08:31:03.599803 Graph has 1 connected components after reconnecting
2023-09-25 08:31:03.600527 68.3% links trimmed from local pruning relative to start
2023-09-25 08:31:03.600550 81.0% links trimmed from global pruning relative to start
2023-09-25 08:31:03.600584 Additional Visual cluster graph pruning for edge bundling at level: 0.15
2023-09-25 08:31:03.602326 Make via clustergraph edgebundle
/home/user/anaconda3/envs/Via2Env_py10/lib/python3.10/site-packages/scipy/sparse/_index.py:103: SparseEfficiencyWarning: Changing the sparsity structure of a csr_matrix is expensive. lil_matrix is more efficient.
self._set_intXint(row, col, x.flat[0])
2023-09-25 08:31:06.258257 Hammer dims: Nodes shape: (146, 2) Edges shape: (918, 3)
2023-09-25 08:31:06.490458 Time elapsed 151.8 seconds
Visualise Via 2.0 Cluster Graph
print(f'{datetime.now()}\t plotting piechart_graph')
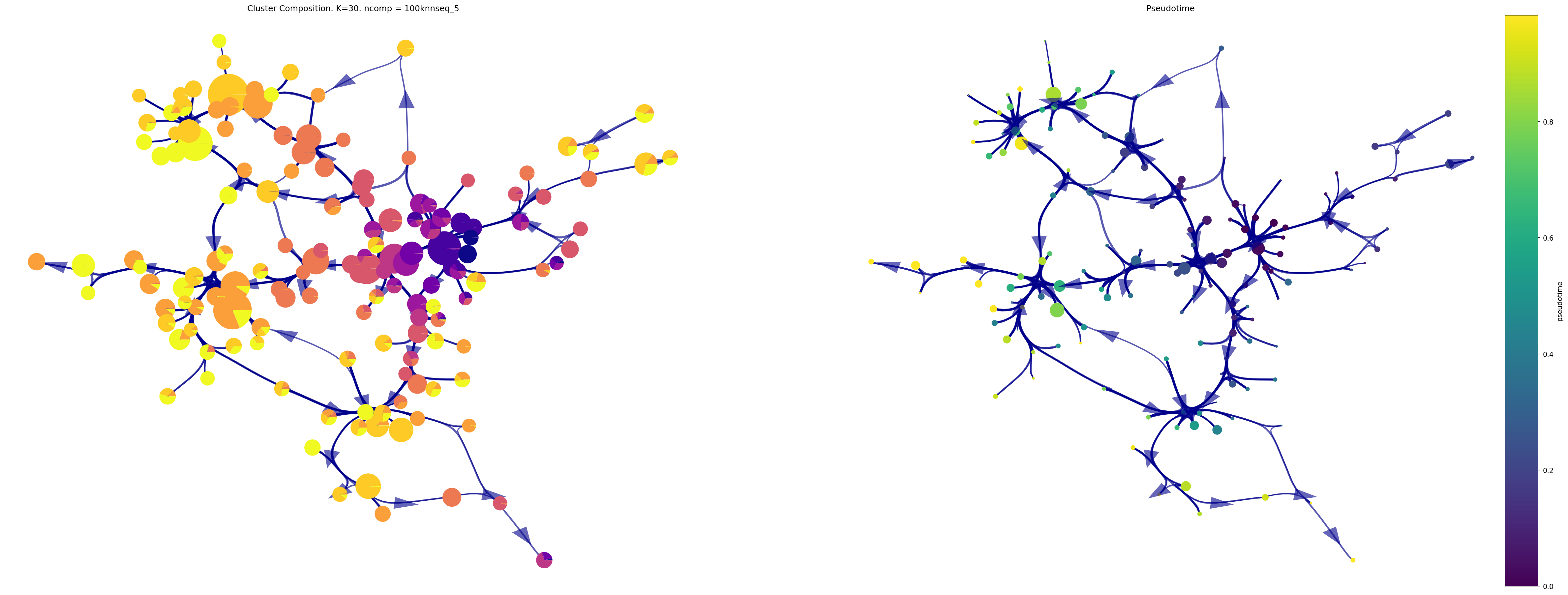
fig,ax1, ax2 = via.draw_piechart_graph(via_object=v0, reference_labels=time_point_numeric, show_legend=False, ax_text=False,
headwidth_arrow=0.8, highlight_terminal_clusters=False, cmap_piechart='plasma', cmap='viridis',
pie_size_scale=0.6,size_node_notpiechart=1)
fig.set_size_inches(40, 15)
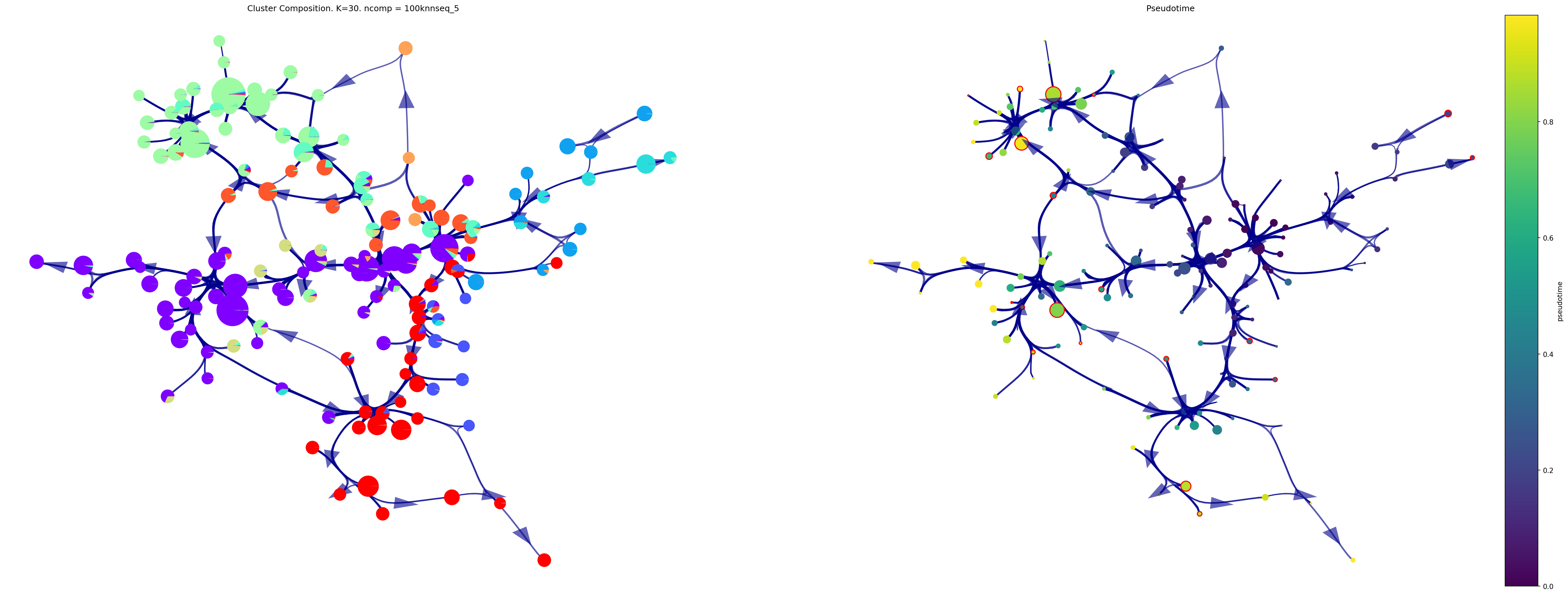
fig,ax1, ax2=via.draw_piechart_graph(via_object=v0, reference_labels=coarse_cell_type, show_legend=False, ax_text=False,
headwidth_arrow=0.8, highlight_terminal_clusters=True, cmap_piechart='rainbow',cmap='viridis',
pie_size_scale=0.5,size_node_notpiechart=1)
fig.set_size_inches(40, 15)
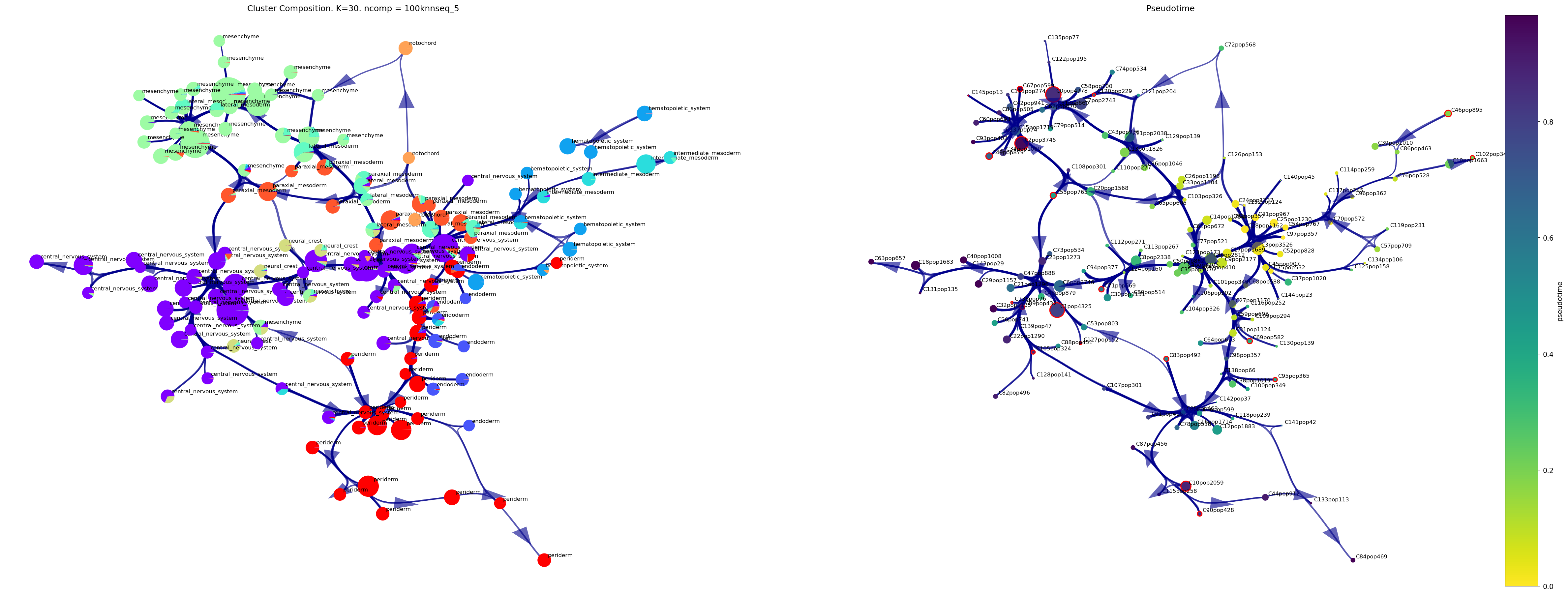
fig,ax1, ax2=via.draw_piechart_graph(via_object=v0, reference_labels=coarse_cell_type, show_legend=False, ax_text=True,
headwidth_arrow=0.8, highlight_terminal_clusters=True, cmap_piechart='rainbow',
pie_size_scale=0.5,size_node_notpiechart=1)
fig.set_size_inches(40, 15)
plt.show()
2023-09-25 08:31:06.497122 plotting piechart_graph
/home/user/PycharmProjects/Via2_May2023_py310/plotting_via.py:3242: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
sct = ax.scatter(node_pos[:, 0], node_pos[:, 1],
/home/user/PycharmProjects/Via2_May2023_py310/plotting_via.py:3242: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
sct = ax.scatter(node_pos[:, 0], node_pos[:, 1],
/home/user/PycharmProjects/Via2_May2023_py310/plotting_via.py:3242: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
sct = ax.scatter(node_pos[:, 0], node_pos[:, 1],
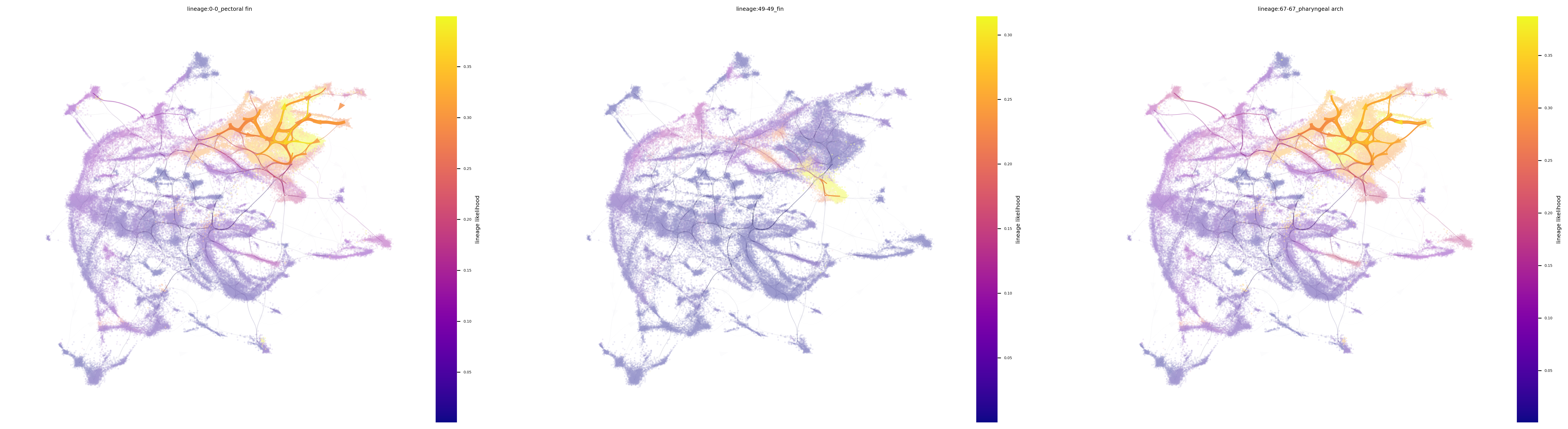
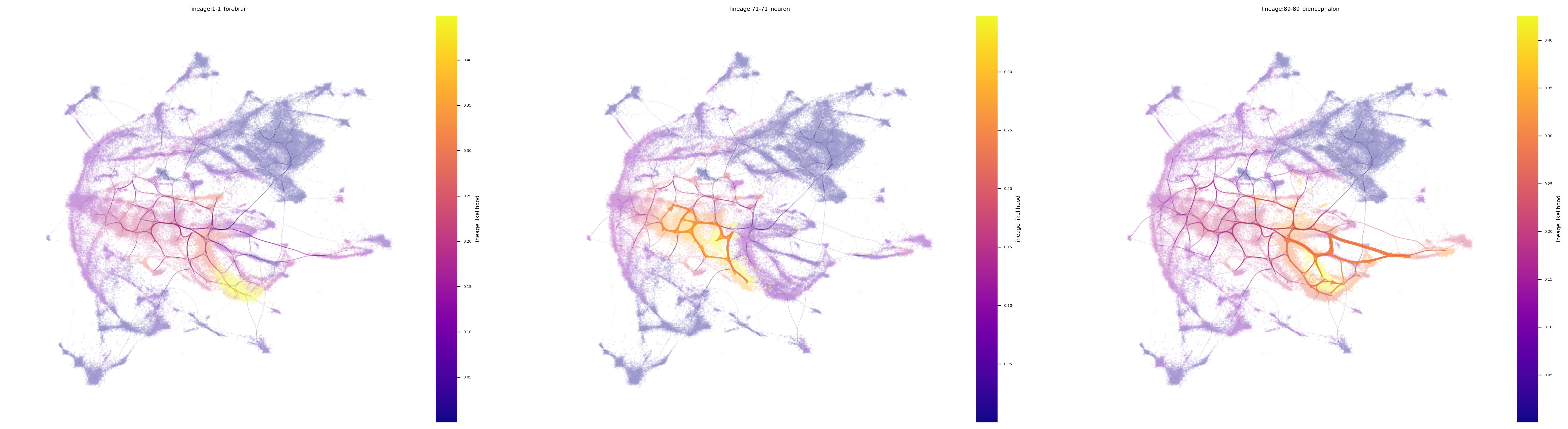
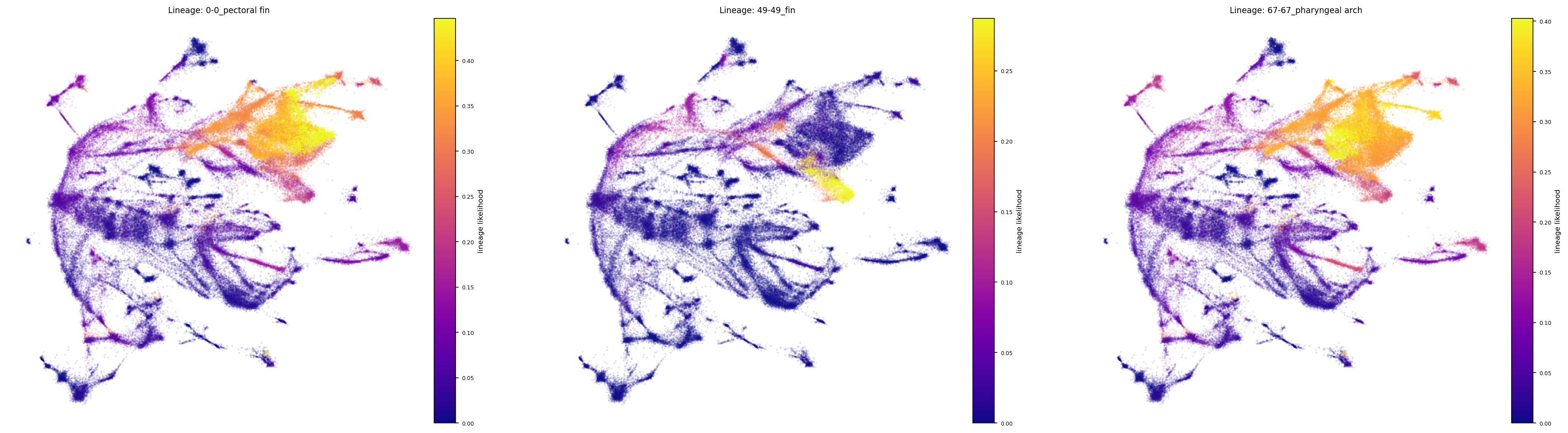
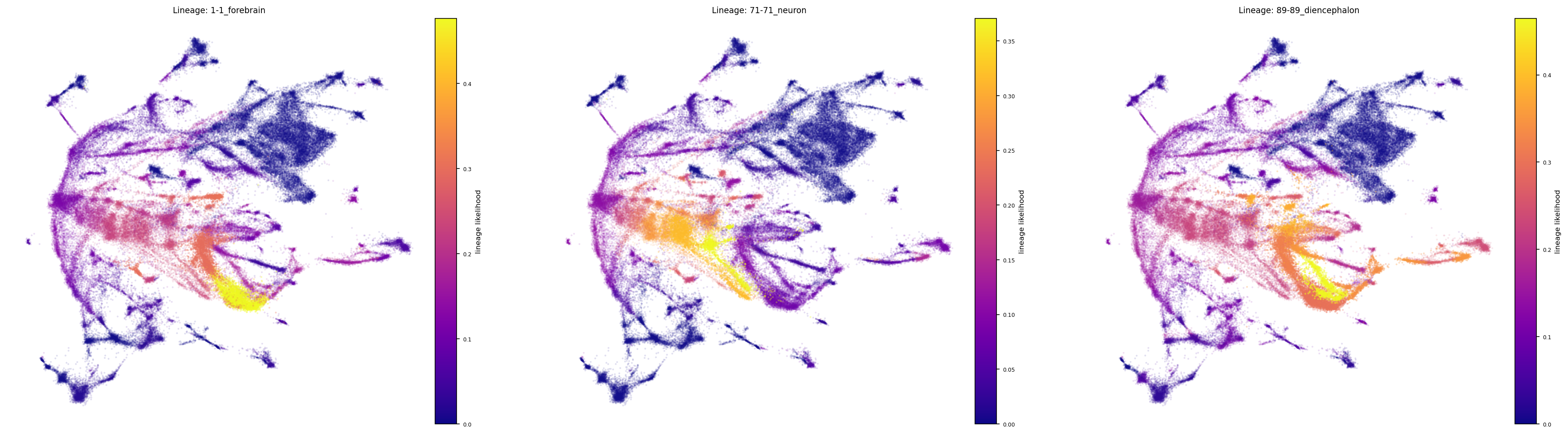
Plot single-cell lineage probabilities
Memory
The lineage probability pathways below are computed based on a memory parameter value of 10 (set when we initialized Via), this is a high degree of memory. The drawback of too high memory can be that some populations are difficult to reach depending on the connectivity of the network. You can always try lowering the value to e.g. 10 or 50.
If you run Via at memory=1 (the no memory case), and then plot the lineage probability pathways and gene trends, you will notice that many of the pathways are diffuse (migrating into unrelated cell populations) and the gene trends can overlap and not be very discriminative.
ecto_lineages=[1,71,89]#,136,10]#,83,115] #CNS
meso_lineages = [0,49,67]#,120,145]#,55,37,46,102]
fig, ax = via.plot_atlas_view(via_object=v0, lineage_pathway=meso_lineages,
linewidth_bundle=3, alpha_bundle_factor=2, headwidth_bundle=0.3,
size_scatter=0.1, alpha_scatter=0.5, use_sc_labels_sequential_for_direction=True)
fig.set_size_inches(30 , 8)
fig, ax = via.plot_atlas_view(via_object=v0, lineage_pathway=ecto_lineages,
linewidth_bundle=3, alpha_bundle_factor=2, headwidth_bundle=0.3,
size_scatter=0.1, alpha_scatter=0.5, use_sc_labels_sequential_for_direction=True)
fig.set_size_inches(30 , 8)
fig, ax = via.draw_sc_lineage_probability(via_object=v0, alpha_factor=0.6, scatter_size=4,
marker_lineages=meso_lineages)
fig.set_size_inches(30 , 8)
fig, ax = via.draw_sc_lineage_probability(via_object=v0, alpha_factor=0.6, scatter_size=4,
marker_lineages=ecto_lineages)
fig.set_size_inches(30 , 8)
plt.show()
2023-09-25 09:15:19.086314 Computing Edges
2023-09-25 09:15:19.086621 Start finding milestones
/home/user/anaconda3/envs/Via2Env_py10/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
2023-09-25 09:15:49.264351 End milestones with 150
2023-09-25 09:15:50.147607 Recompute weights
2023-09-25 09:15:50.186651 pruning milestone graph based on recomputed weights
2023-09-25 09:15:50.188110 Graph has 1 connected components before pruning
2023-09-25 09:15:50.188877 Graph has 1 connected components after pruning
2023-09-25 09:15:50.189092 Graph has 1 connected components after reconnecting
2023-09-25 09:15:50.191130 regenerate igraph on pruned edges
2023-09-25 09:15:50.266700 Setting numeric label as time_series_labels or other sequential metadata for coloring edges
2023-09-25 09:15:50.554701 Making smooth edges
location of 0 is at [0] and 0
location of 49 is at [2] and 2
location of 67 is at [3] and 3
2023-09-25 09:16:25.367232 Computing Edges
2023-09-25 09:16:25.367304 Start finding milestones
/home/user/anaconda3/envs/Via2Env_py10/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
2023-09-25 09:16:53.996278 End milestones with 150
2023-09-25 09:16:54.899173 Recompute weights
2023-09-25 09:16:54.939138 pruning milestone graph based on recomputed weights
2023-09-25 09:16:54.940593 Graph has 1 connected components before pruning
2023-09-25 09:16:54.941371 Graph has 1 connected components after pruning
2023-09-25 09:16:54.941591 Graph has 1 connected components after reconnecting
2023-09-25 09:16:54.943708 regenerate igraph on pruned edges
2023-09-25 09:16:55.014257 Setting numeric label as time_series_labels or other sequential metadata for coloring edges
2023-09-25 09:16:55.267735 Making smooth edges
location of 1 is at [5] and 5
location of 71 is at [6] and 6
location of 89 is at [7] and 7
2023-09-25 09:17:28.852338 Marker_lineages: [0, 49, 67]
2023-09-25 09:17:28.852410 Automatically setting embedding to via_object.embedding
2023-09-25 09:17:28.966372 The number of components in the original full graph is 1
2023-09-25 09:17:28.966428 For downstream visualization purposes we are also constructing a low knn-graph
2023-09-25 09:18:33.313344 Check sc pb 1.0
f getting majority comp
2023-09-25 09:18:39.231618 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 0: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 11, 58, 0]
2023-09-25 09:18:39.231657 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 2: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 43, 79, 15, 34, 2]
2023-09-25 09:18:39.231674 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 49: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 43, 79, 137, 49]
2023-09-25 09:18:39.231688 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 67: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 11, 58, 7, 67]
2023-09-25 09:18:39.231702 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 120: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 11, 58, 0, 120]
2023-09-25 09:18:39.231713 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 1: [97, 3, 17, 61, 113, 80, 53, 1]
2023-09-25 09:18:39.231724 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 71: [97, 3, 17, 61, 113, 80, 53, 1, 71]
2023-09-25 09:18:39.231736 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 89: [97, 3, 17, 9, 92, 50, 8, 23, 32, 89]
2023-09-25 09:18:39.231748 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 136: [97, 3, 17, 61, 113, 80, 53, 1, 6, 48, 136]
2023-09-25 09:18:39.231761 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 10: [97, 3, 52, 45, 68, 27, 59, 31, 109, 95, 141, 44, 90, 10]
2023-09-25 09:18:39.231772 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 83: [97, 3, 52, 45, 68, 27, 59, 31, 83]
2023-09-25 09:18:39.231785 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 145: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 43, 79, 15, 145]
2023-09-25 09:18:39.231796 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 55: [97, 25, 41, 24, 14, 65, 20, 55]
2023-09-25 09:18:39.231806 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 46: [97, 54, 70, 117, 86, 46]
2023-09-25 09:18:39.231815 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 102: [97, 54, 70, 96, 76, 102]
2023-09-25 09:18:39.231826 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 127: [97, 3, 17, 9, 92, 50, 8, 23, 32, 22, 127]
2023-09-25 09:18:39.231839 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 105: [97, 3, 17, 9, 92, 50, 8, 23, 32, 22, 128, 107, 105]
2023-09-25 09:18:39.231851 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 69: [97, 3, 52, 45, 68, 27, 59, 31, 109, 130, 69]
2023-09-25 09:18:39.231864 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 90: [97, 3, 52, 45, 68, 27, 59, 31, 109, 95, 141, 44, 90]
2023-09-25 09:18:39.231875 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 95: [97, 3, 52, 45, 68, 27, 59, 31, 109, 95]
2023-09-25 09:18:39.357220 Revised Cluster level path on sc-knnGraph from Root Cluster 97 to Terminal Cluster 0 along path: [54, 3, 53, 0, 0, 0, 0, 0, 0]
2023-09-25 09:18:39.440505 Revised Cluster level path on sc-knnGraph from Root Cluster 97 to Terminal Cluster 49 along path: [54, 54, 54, 75, 120, 49, 49, 49, 49]
2023-09-25 09:18:39.536499 Revised Cluster level path on sc-knnGraph from Root Cluster 97 to Terminal Cluster 67 along path: [54, 54, 54, 54, 54, 28, 7, 7, 7, 7]
2023-09-25 09:18:39.552106 Marker_lineages: [1, 71, 89]
2023-09-25 09:18:39.552142 Automatically setting embedding to via_object.embedding
2023-09-25 09:18:39.666859 The number of components in the original full graph is 1
2023-09-25 09:18:39.666929 For downstream visualization purposes we are also constructing a low knn-graph
2023-09-25 09:19:43.632672 Check sc pb 1.0
f getting majority comp
2023-09-25 09:19:49.565437 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 0: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 11, 58, 0]
2023-09-25 09:19:49.565473 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 2: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 43, 79, 15, 34, 2]
2023-09-25 09:19:49.565489 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 49: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 43, 79, 137, 49]
2023-09-25 09:19:49.565504 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 67: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 11, 58, 7, 67]
2023-09-25 09:19:49.565518 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 120: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 11, 58, 0, 120]
2023-09-25 09:19:49.565529 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 1: [97, 3, 17, 61, 113, 80, 53, 1]
2023-09-25 09:19:49.565540 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 71: [97, 3, 17, 61, 113, 80, 53, 1, 71]
2023-09-25 09:19:49.565553 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 89: [97, 3, 17, 9, 92, 50, 8, 23, 32, 89]
2023-09-25 09:19:49.565569 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 136: [97, 3, 17, 61, 113, 80, 53, 1, 6, 48, 136]
2023-09-25 09:19:49.565583 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 10: [97, 3, 52, 45, 68, 27, 59, 31, 109, 95, 141, 44, 90, 10]
2023-09-25 09:19:49.565594 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 83: [97, 3, 52, 45, 68, 27, 59, 31, 83]
2023-09-25 09:19:49.565606 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 145: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 43, 79, 15, 145]
2023-09-25 09:19:49.565617 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 55: [97, 25, 41, 24, 14, 65, 20, 55]
2023-09-25 09:19:49.565627 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 46: [97, 54, 70, 117, 86, 46]
2023-09-25 09:19:49.565637 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 102: [97, 54, 70, 96, 76, 102]
2023-09-25 09:19:49.565648 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 127: [97, 3, 17, 9, 92, 50, 8, 23, 32, 22, 127]
2023-09-25 09:19:49.565661 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 105: [97, 3, 17, 9, 92, 50, 8, 23, 32, 22, 128, 107, 105]
2023-09-25 09:19:49.565673 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 69: [97, 3, 52, 45, 68, 27, 59, 31, 109, 130, 69]
2023-09-25 09:19:49.565685 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 90: [97, 3, 52, 45, 68, 27, 59, 31, 109, 95, 141, 44, 90]
2023-09-25 09:19:49.565696 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 95: [97, 3, 52, 45, 68, 27, 59, 31, 109, 95]
2023-09-25 09:19:49.689435 Revised Cluster level path on sc-knnGraph from Root Cluster 97 to Terminal Cluster 1 along path: [54, 3, 1, 1, 1, 1]
2023-09-25 09:19:49.777811 Revised Cluster level path on sc-knnGraph from Root Cluster 97 to Terminal Cluster 71 along path: [54, 3, 80, 71, 71]
2023-09-25 09:19:49.872623 Revised Cluster level path on sc-knnGraph from Root Cluster 97 to Terminal Cluster 89 along path: [54, 3, 1, 89]
Differentiation Flow
Generates a differentiation flow diagram based on the Via2.0 clustergraph. The second plot in each function call has the details by parc cluster and major cell population. In this case, we assign a different root node for each set of lineages.
ecto_lineages=[1,22,71,89,136,10,83,115] #CNS
meso_lineages = [0,49,67,120,145,55,37,46,102]
via.draw_differentiation_flow(via_object=v0, marker_lineages=meso_lineages, label_node=coarse_fine_cell_type,
do_log_flow=True, root_cluster_list=[97]) # cluster corresponding to root
via.draw_differentiation_flow(via_object=v0, marker_lineages=ecto_lineages, label_node=coarse_fine_cell_type,
do_log_flow=True, root_cluster_list=[52]) # cluster corresponding to a suitable CNS root
plt.show()
2023-09-25 08:49:15.613940 Marker_lineages: [0, 49, 67, 120, 145, 55, 37, 46, 102]
automatically setting embedding to via_object.embedding
2023-09-25 08:49:15.614169 Start dictionary modes
2023-09-25 08:49:15.852553 End dictionary modes
2023-09-25 08:49:18.016249 Check sc pb 1.0
2023-09-25 08:49:25.668206 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 0: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 11, 58, 0]
2023-09-25 08:49:25.741354 Holoviews for TC 0
dest 0, is at loc 0 on the bp_array
2023-09-25 08:49:25.741804 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 49: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 43, 79, 137, 49]
2023-09-25 08:49:25.806408 Holoviews for TC 49
dest 49, is at loc 2 on the bp_array
2023-09-25 08:49:25.807038 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 67: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 11, 58, 7, 67]
2023-09-25 08:49:25.840009 Holoviews for TC 67
dest 67, is at loc 3 on the bp_array
2023-09-25 08:49:25.840436 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 120: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 11, 58, 0, 120]
2023-09-25 08:49:25.889504 Holoviews for TC 120
dest 120, is at loc 4 on the bp_array
2023-09-25 08:49:25.889883 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 145: [97, 25, 41, 24, 14, 65, 20, 110, 36, 26, 43, 79, 15, 145]
2023-09-25 08:49:25.941053 Holoviews for TC 145
dest 145, is at loc 11 on the bp_array
2023-09-25 08:49:25.941432 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 55: [97, 25, 41, 24, 14, 65, 20, 55]
2023-09-25 08:49:26.001078 Holoviews for TC 55
dest 55, is at loc 12 on the bp_array
2023-09-25 08:49:26.001825 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 37: [97, 54, 70, 57, 125, 37]
2023-09-25 08:49:26.052467 Holoviews for TC 37
2023-09-25 08:49:26.052785 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 46: [97, 54, 70, 117, 86, 46]
2023-09-25 08:49:26.113529 Holoviews for TC 46
dest 46, is at loc 13 on the bp_array
2023-09-25 08:49:26.114352 Cluster path on clustergraph starting from Root Cluster 97 to Terminal Cluster 102: [97, 54, 70, 96, 76, 102]
2023-09-25 08:49:26.179086 Holoviews for TC 102
dest 102, is at loc 14 on the bp_array
2023-09-25 08:49:26.182116 Start sankey
2023-09-25 08:49:26.272915 make sankey color dict
2023-09-25 08:49:26.274535 set options and render
2023-09-25 08:49:27.354921 Marker_lineages: [1, 22, 71, 89, 136, 10, 83, 115]
automatically setting embedding to via_object.embedding
2023-09-25 08:49:27.355000 Start dictionary modes
2023-09-25 08:49:27.545101 End dictionary modes
2023-09-25 08:49:29.465100 Check sc pb 1.0
2023-09-25 08:49:35.972127 Cluster path on clustergraph starting from Root Cluster 52 to Terminal Cluster 1: [52, 3, 17, 61, 113, 80, 53, 1]
2023-09-25 08:49:36.004311 Holoviews for TC 1
dest 1, is at loc 5 on the bp_array
2023-09-25 08:49:36.004850 Cluster path on clustergraph starting from Root Cluster 52 to Terminal Cluster 22: [52, 3, 17, 9, 92, 50, 8, 23, 32, 22]
2023-09-25 08:49:36.043009 Holoviews for TC 22
2023-09-25 08:49:36.043484 Cluster path on clustergraph starting from Root Cluster 52 to Terminal Cluster 71: [52, 3, 17, 61, 113, 80, 53, 1, 71]
2023-09-25 08:49:36.088468 Holoviews for TC 71
dest 71, is at loc 6 on the bp_array
2023-09-25 08:49:36.089030 Cluster path on clustergraph starting from Root Cluster 52 to Terminal Cluster 89: [52, 3, 17, 9, 92, 50, 8, 23, 32, 89]
2023-09-25 08:49:36.139421 Holoviews for TC 89
dest 89, is at loc 7 on the bp_array
2023-09-25 08:49:36.139808 Cluster path on clustergraph starting from Root Cluster 52 to Terminal Cluster 136: [52, 3, 17, 61, 113, 80, 53, 1, 6, 48, 136]
2023-09-25 08:49:36.198432 Holoviews for TC 136
dest 136, is at loc 8 on the bp_array
2023-09-25 08:49:36.198699 Cluster path on clustergraph starting from Root Cluster 52 to Terminal Cluster 10: [52, 45, 68, 27, 59, 31, 109, 95, 141, 44, 90, 10]
2023-09-25 08:49:36.264525 Holoviews for TC 10
dest 10, is at loc 9 on the bp_array
2023-09-25 08:49:36.264947 Cluster path on clustergraph starting from Root Cluster 52 to Terminal Cluster 83: [52, 45, 68, 27, 59, 31, 83]
2023-09-25 08:49:36.322102 Holoviews for TC 83
dest 83, is at loc 10 on the bp_array
2023-09-25 08:49:36.322475 Cluster path on clustergraph starting from Root Cluster 52 to Terminal Cluster 115: [52, 45, 68, 27, 98, 138, 142, 66, 78, 115]
2023-09-25 08:49:36.379335 Holoviews for TC 115
2023-09-25 08:49:36.381979 Start sankey
2023-09-25 08:49:36.459963 make sankey color dict
2023-09-25 08:49:36.462874 set options and render